Genetic associations between gut microbiota and allergic rhinitis: an LDSC and MR analysis
- PMID: 38855765
- PMCID: PMC11157438
- DOI: 10.3389/fmicb.2024.1395340
Genetic associations between gut microbiota and allergic rhinitis: an LDSC and MR analysis
Abstract
Background: Several studies have suggested a potential link between allergic rhinitis (AR) and gut microbiota. In response, we conducted a meta-analysis of Linkage Disequilibrium Score Regression (LDSC) and Mendelian randomization (MR) to detect their genetic associations.
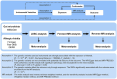
Methods: Summary statistics for 211 gut microbiota taxa were gathered from the MiBioGen study, while data for AR were sourced from the Pan-UKB, the FinnGen, and the Genetic Epidemiology Research on Aging (GERA). The genetic correlation between gut microbiota and AR was assessed using LDSC. The principal estimate of causality was determined using the Inverse-Variance Weighted (IVW) method. To assess the robustness of these findings, sensitivity analyses were conducted employing methods such as the weighted median, MR-Egger, and MR-PRESSO. The summary effect estimates of LDSC, forward MR and reverse MR were combined using meta-analysis for AR from different data resources.
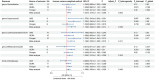
Results: Our study indicated a significant genetic correlation between genus Sellimonas (Rg = -0.64, p = 3.64 × 10-5, Adjust_P = 3.64 × 10-5) and AR, and a suggestive genetic correlation between seven bacterial taxa and AR. Moreover, the forward MR analysis identified genus Gordonibacter, genus Coprococcus2, genus LachnospiraceaeUCG010, genus Methanobrevibacter, and family Victivallaceae as being suggestively associated with an increased risk of AR. The reverse MR analysis indicated that AR was suggestively linked to an increased risk for genus Coprococcus2 and genus RuminococcaceaeUCG011.
Conclusion: Our findings indicate a causal relationship between specific gut microbiomes and AR. This enhances our understanding of the gut microbiota's contribution to the pathophysiology of AR and lays the groundwork for innovative approaches and theoretical models for future prevention and treatment strategies in this patient population.
Keywords: Mendelian randomization; allergic rhinitis; gut microbiota; linkage disequilibrium score regression; meta-analysis.
Copyright © 2024 Zheng, Chen, Zhuang, Xu, Zhao, Qian and Shen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Genetic associations between gut microbiota and type 2 diabetes mediated by plasma metabolites: a Mendelian randomization study.Front Endocrinol (Lausanne). 2024 Aug 9;15:1430675. doi: 10.3389/fendo.2024.1430675. eCollection 2024. Front Endocrinol (Lausanne). 2024. PMID: 39184139 Free PMC article.
-
Mendelian randomization supports causality between gut microbiota and chronic hepatitis B.Front Microbiol. 2023 Aug 16;14:1243811. doi: 10.3389/fmicb.2023.1243811. eCollection 2023. Front Microbiol. 2023. PMID: 37655340 Free PMC article.
-
Unveiling genetic links between gut microbiota and asthma: a Mendelian randomization.Front Microbiol. 2024 Sep 20;15:1448629. doi: 10.3389/fmicb.2024.1448629. eCollection 2024. Front Microbiol. 2024. PMID: 39372270 Free PMC article.
-
Genetic liability of gut microbiota for idiopathic pulmonary fibrosis and lung function: a two-sample Mendelian randomization study.Front Cell Infect Microbiol. 2024 May 22;14:1348685. doi: 10.3389/fcimb.2024.1348685. eCollection 2024. Front Cell Infect Microbiol. 2024. PMID: 38841114 Free PMC article.
-
Exploring the potential causal relationship between gut microbiota and heart failure: A two-sample mendelian randomization study combined with the geo database.Curr Probl Cardiol. 2024 Feb;49(2):102235. doi: 10.1016/j.cpcardiol.2023.102235. Epub 2023 Nov 30. Curr Probl Cardiol. 2024. PMID: 38040216 Review.
Cited by
-
Exploring the relationship between gut microbiota, immune characteristics, and female genital tract polyps using genetic evidence.Medicine (Baltimore). 2024 Dec 6;103(49):e40833. doi: 10.1097/MD.0000000000040833. Medicine (Baltimore). 2024. PMID: 39654186 Free PMC article.
References
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B 57, 289–300. doi: 10.1111/j.2517-6161.1995.tb02031.x - DOI
-
- Bibbò S., Ianiro G., Giorgio V., Scaldaferri F., Masucci L., Gasbarrini A., et al. . (2016). The role of diet on gut microbiota composition. Eur. Rev. Med. Pharmacol. Sci. 20, 4742–4749, PMID: - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

