Use of a diagnostic Puumala virus real-time RT-PCR in an orthohantavirus endemic region in the Netherlands
- PMID: 38856680
- PMCID: PMC11218528
- DOI: 10.1128/spectrum.03813-23
Use of a diagnostic Puumala virus real-time RT-PCR in an orthohantavirus endemic region in the Netherlands
Abstract
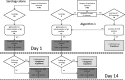
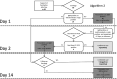
Laboratory diagnosis of orthohantavirus infection is primarily based on serology. However, for a confirmed serological diagnosis, evaluation of a follow-up serum sample is essential, which is time consuming and causes delay. Real-time reverse transcription polymerase chain reaction (RT-PCR) tests, if positive, provide an immediate and definitive diagnosis, and accurately identify the causative agent, where the discriminative nature of serology is suboptimal. We re-evaluated sera from orthohantavirus-suspected clinical cases in the Dutch regions of Twente and Achterhoek from July 2014 to April 2016 for the presence of Puumala orthohantavirus (PUUV), Tula orthohantavirus (TULV), and Seoul orthohantavirus (SEOV) RNA. PUUV RNA was detected in 11% of the total number (n = 85) of sera tested, in 50% of sera positive for anti-PUUV/TULV IgM (n = 16), and in 1.4% of sera negative or indeterminate for anti-PUUV/TULV IgM (n = 69). No evidence was found for the presence of TULV or SEOV viral RNA. Based on these findings, we propose two algorithms to implement real-time RT-PCR testing in routine orthohantavirus diagnostics, which optimally provide clinicians with early confirmed diagnoses and could prevent possible further invasive testing and treatment.
Importance: The addition of a real-time reverse transcription polymerase chain reaction test to routine orthohantavirus diagnostics may better aid clinical decision making than the use of standard serology tests alone. Awareness by clinicians and clinical microbiologists of this advantage may ultimately lead to a reduction in over-hospitalization and unnecessary invasive diagnostic procedures.
Keywords: Puumala virus; diagnostics; hantavirus; molecular methods; nephrology; nucleic acid amplification test; serology; zoonotic infections.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Molecular and epidemiological characteristics of human Puumala and Dobrava-Belgrade hantavirus infections, Germany, 2001 to 2017.Euro Surveill. 2019 Aug;24(32):1800675. doi: 10.2807/1560-7917.ES.2019.24.32.1800675. Euro Surveill. 2019. PMID: 31411134 Free PMC article.
-
Puumala hantavirus viremia diagnosed by real-time reverse transcriptase PCR using samples from patients with hemorrhagic fever and renal syndrome.J Clin Microbiol. 2007 Aug;45(8):2491-7. doi: 10.1128/JCM.01902-06. Epub 2007 May 30. J Clin Microbiol. 2007. PMID: 17537944 Free PMC article.
-
Development of a Comparative European Orthohantavirus Microneutralization Assay With Multi- Species Validation and Evaluation in a Human Diagnostic Cohort.Front Cell Infect Microbiol. 2020 Dec 22;10:580478. doi: 10.3389/fcimb.2020.580478. eCollection 2020. Front Cell Infect Microbiol. 2020. PMID: 33415084 Free PMC article.
-
Seoul hantavirus in brown rats in the Netherlands: implications for physicians--Epidemiology, clinical aspects, treatment and diagnostics.Neth J Med. 2015 May;73(4):155-60. Neth J Med. 2015. PMID: 25968286 Review.
-
A Review of Hantavirus Research in Indonesia: Prevalence in Humans and Rodents, and the Discovery of Serang Virus.Viruses. 2019 Jul 31;11(8):698. doi: 10.3390/v11080698. Viruses. 2019. PMID: 31370291 Free PMC article. Review.
Cited by
-
Orthohantaviruses: An Overview of the Current Status of Diagnostics and Surveillance.Viruses. 2025 Apr 26;17(5):622. doi: 10.3390/v17050622. Viruses. 2025. PMID: 40431633 Free PMC article. Review.
-
Puumala orthohantavirus: prevalence, biology, disease, animal models and recent advances in therapeutics development and structural biology.Front Immunol. 2025 May 8;16:1575112. doi: 10.3389/fimmu.2025.1575112. eCollection 2025. Front Immunol. 2025. PMID: 40406115 Free PMC article. Review.
References
-
- Adams MJ, Lefkowitz EJ, King AMQ, Harrach B, Harrison RL, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Mushegian AR, Nibert M, Sabanadzovic S, Sanfaçon H, Siddell SG, Simmonds P, Varsani A, Zerbini FM, Gorbalenya AE, Davison AJ. 2017. Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2017). Arch Virol 162:2505–2538. doi:10.1007/s00705-017-3358-5 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

