Characterization of organelle DNA degradation mediated by DPD1 exonuclease in the rice genome-edited line
- PMID: 38856917
- PMCID: PMC11164812
- DOI: 10.1007/s11103-024-01452-x
Characterization of organelle DNA degradation mediated by DPD1 exonuclease in the rice genome-edited line
Abstract
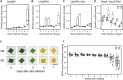
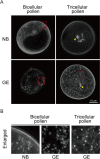
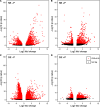
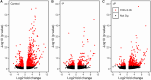
Mitochondria and plastids, originated as ancestral endosymbiotic bacteria, contain their own DNA sequences. These organelle DNAs (orgDNAs) are, despite the limited genetic information they contain, an indispensable part of the genetic systems but exist as multiple copies, making up a substantial amount of total cellular DNA. Given this abundance, orgDNA is known to undergo tissue-specific degradation in plants. Previous studies have shown that the exonuclease DPD1, conserved among seed plants, degrades orgDNAs during pollen maturation and leaf senescence in Arabidopsis. However, tissue-specific orgDNA degradation was shown to differ among species. To extend our knowledge, we characterized DPD1 in rice in this study. We created a genome-edited (GE) mutant in which OsDPD1 and OsDPD1-like were inactivated. Characterization of this GE plant demonstrated that DPD1 was involved in pollen orgDNA degradation, whereas it had no significant effect on orgDNA degradation during leaf senescence. Comparison of transcriptomes from wild-type and GE plants with different phosphate supply levels indicated that orgDNA had little impact on the phosphate starvation response, but instead had a global impact in plant growth. In fact, the GE plant showed lower fitness with reduced grain filling rate and grain weight in natural light conditions. Taken together, the presented data reinforce the important physiological roles of orgDNA degradation mediated by DPD1.
Keywords: Exonuclease; Leaf senescence; Mitochondria; Organelle DNA degradation; Plastids; Pollen; Rice (Oryza sativa).
© 2024. The Author(s).
Conflict of interest statement
The authors have no conflicts of interest to declare.
Figures



References
-
- Corriveau JL, Coleman AW. Rapid screening method to detect potential biparental inheritance of plastid DNA and results fo over 200 angiosperm species. Am J Bot. 1988;75:1443–1458. doi: 10.1002/j.1537-2197.1988.tb11219.x. - DOI
-
- Dyall SD, Brown MT, Johnson PJ. Ancient invasions: from endosymbionts to Organelles. Sci New Ser. 2004;304(5668):253–257. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

