A Bibliometric Analysis of the Spatial Transcriptomics Literature from 2006 to 2023
- PMID: 38856921
- PMCID: PMC11164738
- DOI: 10.1007/s10571-024-01484-3
A Bibliometric Analysis of the Spatial Transcriptomics Literature from 2006 to 2023
Abstract
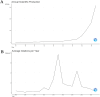
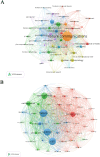
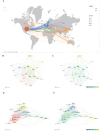
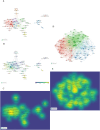
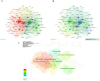
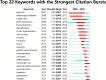
In recent years, spatial transcriptomics (ST) research has become a popular field of study and has shown great potential in medicine. However, there are few bibliometric analyses in this field. Thus, in this study, we aimed to find and analyze the frontiers and trends of this medical research field based on the available literature. A computerized search was applied to the WoSCC (Web of Science Core Collection) Database for literature published from 2006 to 2023. Complete records of all literature and cited references were extracted and screened. The bibliometric analysis and visualization were performed using CiteSpace, VOSviewer, Bibliometrix R Package software, and Scimago Graphica. A total of 1467 papers and reviews were included. The analysis revealed that the ST publication and citation results have shown a rapid upward trend over the last 3 years. Nature Communications and Nature were the most productive and most co-cited journals, respectively. In the comprehensive global collaborative network, the United States is the country with the most organizations and publications, followed closely by China and the United Kingdom. The author Joakim Lundeberg published the most cited paper, while Patrik L. Ståhl ranked first among co-cited authors. The hot topics in ST are tissue recognition, cancer, heterogeneity, immunotherapy, differentiation, and models. ST technologies have greatly contributed to in-depth research in medical fields such as oncology and neuroscience, opening up new possibilities for the diagnosis and treatment of diseases. Moreover, artificial intelligence and big data drive additional development in ST fields.
Keywords: Bibliometric analysis; Cancer; Differentiation; Models; Spatial transcriptomics; Tissue recognition.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Research Trends in the Application of Artificial Intelligence in Oncology: A Bibliometric and Network Visualization Study.Front Biosci (Landmark Ed). 2022 Aug 31;27(9):254. doi: 10.31083/j.fbl2709254. Front Biosci (Landmark Ed). 2022. PMID: 36224012
-
Worldwide productivity and research trend of publications concerning tumor immune microenvironment (TIME): a bibliometric study.Eur J Med Res. 2023 Jul 10;28(1):229. doi: 10.1186/s40001-023-01195-3. Eur J Med Res. 2023. PMID: 37430294 Free PMC article.
-
Knowledge Structure and Emerging Trends of Telerehabilitation in Recent 20 Years: A Bibliometric Analysis via CiteSpace.Front Public Health. 2022 Jun 20;10:904855. doi: 10.3389/fpubh.2022.904855. eCollection 2022. Front Public Health. 2022. PMID: 35795695 Free PMC article.
-
Global trends and hotspots of treatment for nonalcoholic fatty liver disease: A bibliometric and visualization analysis (2010-2023).World J Gastroenterol. 2023 Oct 7;29(37):5339-5360. doi: 10.3748/wjg.v29.i37.5339. World J Gastroenterol. 2023. PMID: 37899789 Free PMC article. Review.
-
Bibliometric analysis of bone metastases from lung cancer research from 2004 to 2023.Front Oncol. 2024 Aug 6;14:1439209. doi: 10.3389/fonc.2024.1439209. eCollection 2024. Front Oncol. 2024. PMID: 39165682 Free PMC article.
Cited by
-
Research hotspots and emerging topics in neuromyelitis optica spectrum disorder treatment: Insights from a bibliometric analysis.Medicine (Baltimore). 2025 Jun 6;104(23):e42850. doi: 10.1097/MD.0000000000042850. Medicine (Baltimore). 2025. PMID: 40489809 Free PMC article. Review.
-
spatialGE Is a User-Friendly Web Application That Facilitates Spatial Transcriptomics Data Analysis.Cancer Res. 2025 Mar 3;85(5):848-858. doi: 10.1158/0008-5472.CAN-24-2346. Cancer Res. 2025. PMID: 39636739 Free PMC article.
-
spatialGE: A user-friendly web application to democratize spatial transcriptomics analysis.bioRxiv [Preprint]. 2024 Jul 2:2024.06.27.601050. doi: 10.1101/2024.06.27.601050. bioRxiv. 2024. Update in: Cancer Res. 2025 Mar 03;85(5):848-858. doi: 10.1158/0008-5472.CAN-24-2346. PMID: 39005315 Free PMC article. Updated. Preprint.
References
-
- Al-Sabah J, Baccin C, Haas S (2019) Single-cell and spatial transcriptomics approaches of the bone marrow microenvironment. Curr Opin Oncol 32(2):146–153 - PubMed
-
- Aria M, Cuccurullo C (2017) bibliometrix: an R-tool for comprehensive science mapping analysis. J Informet 11(4):959–975. 10.1016/j.joi.2017.08.007
-
- Bao Z, Wang Y, Wang Q, Fang S, Shan X, Wang J, Jiang T (2021) Intratumor heterogeneity, microenvironment, and mechanisms of drug resistance in glioma recurrence and evolution. Front Med 15(4):551–561. 10.1007/s11684-020-0760-2 - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

