Vertebrate centromere architecture: from chromatin threads to functional structures
- PMID: 38856923
- PMCID: PMC11266386
- DOI: 10.1007/s00412-024-00823-z
Vertebrate centromere architecture: from chromatin threads to functional structures
Abstract
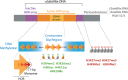
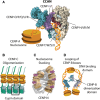
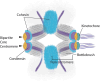
Centromeres are chromatin structures specialized in sister chromatid cohesion, kinetochore assembly, and microtubule attachment during chromosome segregation. The regional centromere of vertebrates consists of long regions of highly repetitive sequences occupied by the Histone H3 variant CENP-A, and which are flanked by pericentromeres. The three-dimensional organization of centromeric chromatin is paramount for its functionality and its ability to withstand spindle forces. Alongside CENP-A, key contributors to the folding of this structure include components of the Constitutive Centromere-Associated Network (CCAN), the protein CENP-B, and condensin and cohesin complexes. Despite its importance, the intricate architecture of the regional centromere of vertebrates remains largely unknown. Recent advancements in long-read sequencing, super-resolution and cryo-electron microscopy, and chromosome conformation capture techniques have significantly improved our understanding of this structure at various levels, from the linear arrangement of centromeric sequences and their epigenetic landscape to their higher-order compaction. In this review, we discuss the latest insights on centromere organization and place them in the context of recent findings describing a bipartite higher-order organization of the centromere.
Keywords: CENP-A; Centromere; Chromatin organization; Epigenetics; Kinetochore.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
References
-
- Altemose N, Logsdon GA, Bzikadze AV, Sidhwani P, Langley SA, Caldas GV, Hoyt SJ, Uralsky L, Ryabov FD, Shew CJ, Sauria MEG, Borchers M, Gershman A, Mikheenko A, Shepelev VA, Dvorkina T, Kunyavskaya O, Vollger MR, Rhie A, … Miga KH (2022a) Complete genomic and epigenetic maps of human centromeres. Science (New York, N.Y.) 376(6588):eabl4178. 10.1126/science.abl4178 - PMC - PubMed
-
- Altemose N, Maslan A, Smith OK, Sundararajan K, Brown RR, Mishra R, Detweiler AM, Neff N, Miga KH, Straight AF, Streets A (2022b) DiMeLo-seq: a long-read, single-molecule method for mapping protein–DNA interactions genome wide. Nat Methods 19(6):711. 10.1038/s41592-022-01475-6 10.1038/s41592-022-01475-6 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

