CHARMM-GUI QM/MM Interfacer for a Quantum Mechanical and Molecular Mechanical (QM/MM) Simulation Setup: 1. Semiempirical Methods
- PMID: 38856971
- PMCID: PMC11209942
- DOI: 10.1021/acs.jctc.4c00439
CHARMM-GUI QM/MM Interfacer for a Quantum Mechanical and Molecular Mechanical (QM/MM) Simulation Setup: 1. Semiempirical Methods
Abstract
Quantum mechanical (QM) treatments, when combined with molecular mechanical (MM) force fields, can effectively handle enzyme-catalyzed reactions without significantly increasing the computational cost. In this context, we present CHARMM-GUI QM/MM Interfacer, a web-based cyberinfrastructure designed to streamline the preparation of various QM/MM simulation inputs with ligand modification. The development of QM/MM Interfacer has been achieved through integration with existing CHARMM-GUI modules, such as PDB Reader and Manipulator, Solution Builder, and Membrane Builder. In addition, new functionalities have been developed to facilitate the one-stop preparation of QM/MM systems and enable interactive and intuitive ligand modifications and QM atom selections. QM/MM Interfacer offers support for a range of semiempirical QM methods, including AM1(+/d), PM3(+/PDDG), MNDO(+/d, +/PDDG), PM6, RM1, and SCC-DFTB, tailored for both AMBER and CHARMM. A nontrivial setup related to ligand modification, link-atom insertion, and charge distribution is automatized through intuitive user interfaces. To illustrate the robustness of QM/MM Interfacer, we conducted QM/MM simulations of three enzyme-substrate systems: dihydrofolate reductase, insulin receptor kinase, and oligosaccharyltransferase. In addition, we have created three tutorial videos about building these systems, which can be found at https://www.charmm-gui.org/demo/qmi. QM/MM Interfacer is expected to be a valuable and accessible web-based tool that simplifies and accelerates the setup process for hybrid QM/MM simulations.
Conflict of interest statement
The authors declare the following competing financial interest(s): W.I. is the co-founder and CEO of MolCube INC.
Figures






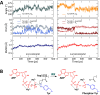
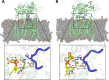

References
-
- Knowles J. R.; Albery W. J. Perfection in enzyme catalysis: the energetics of triosephosphate isomerase. Acc. Chem. Res. 1977, 10, 105–111. 10.1021/ar50112a001. - DOI
-
- Showen R. In Transition states of biochemical processes. Part 1. Gandour R. D., Schowen R. L., Eds.; Plenum Press, New York, 1978.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
