PARP-1 selectively impairs KRAS-driven phenotypic and molecular features in intrahepatic cholangiocarcinoma
- PMID: 38857989
- PMCID: PMC11420749
- DOI: 10.1136/gutjnl-2023-331237
PARP-1 selectively impairs KRAS-driven phenotypic and molecular features in intrahepatic cholangiocarcinoma
Abstract
Objective: Intrahepatic cholangiocarcinoma (iCCA) is the second most common primary liver cancer with limited therapeutic options. KRAS mutations are among the most abundant genetic alterations in iCCA associated with poor clinical outcome and treatment response. Recent findings indicate that Poly(ADP-ribose)polymerase1 (PARP-1) is implicated in KRAS-driven cancers, but its exact role in cholangiocarcinogenesis remains undefined.
Design: PARP-1 inhibition was performed in patient-derived and established iCCA cells using RNAi, CRISPR/Cas9 and pharmacological inhibition in KRAS-mutant, non-mutant cells. In addition, Parp-1 knockout mice were combined with iCCA induction by hydrodynamic tail vein injection to evaluate an impact on phenotypic and molecular features of Kras-driven and Kras-wildtype iCCA. Clinical implications were confirmed in authentic human iCCA.
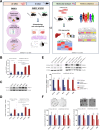
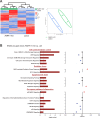
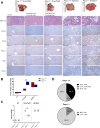
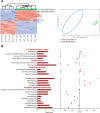
Results: PARP-1 was significantly enhanced in KRAS-mutant human iCCA. PARP-1-based interventions preferentially impaired cell viability and tumourigenicity in human KRAS-mutant cell lines. Consistently, loss of Parp-1 provoked distinct phenotype in Kras/Tp53-induced versus Akt/Nicd-induced iCCA and abolished Kras-dependent cholangiocarcinogenesis. Transcriptome analyses confirmed preferential impairment of DNA damage response pathways and replicative stress response mediated by CHK1. Consistently, inhibition of CHK1 effectively reversed PARP-1 mediated effects. Finally, Parp-1 depletion induced molecular switch of KRAS-mutant iCCA recapitulating good prognostic human iCCA patients.
Conclusion: Our findings identify the novel prognostic and therapeutic role of PARP-1 in iCCA patients with activation of oncogenic KRAS signalling.
Keywords: CARCINOGENESIS; CHOLANGIOCARCINOMA; GENE MUTATION; MOLECULAR ONCOLOGY.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
