Patient-derived organoid biobank identifies epigenetic dysregulation of intestinal epithelial MHC-I as a novel mechanism in severe Crohn's Disease
- PMID: 38857990
- PMCID: PMC11347221
- DOI: 10.1136/gutjnl-2024-332043
Patient-derived organoid biobank identifies epigenetic dysregulation of intestinal epithelial MHC-I as a novel mechanism in severe Crohn's Disease
Abstract
Objective: Epigenetic mechanisms, including DNA methylation (DNAm), have been proposed to play a key role in Crohn's disease (CD) pathogenesis. However, the specific cell types and pathways affected as well as their potential impact on disease phenotype and outcome remain unknown. We set out to investigate the role of intestinal epithelial DNAm in CD pathogenesis.
Design: We generated 312 intestinal epithelial organoids (IEOs) from mucosal biopsies of 168 patients with CD (n=72), UC (n=23) and healthy controls (n=73). We performed genome-wide molecular profiling including DNAm, bulk as well as single-cell RNA sequencing. Organoids were subjected to gene editing and the functional consequences of DNAm changes evaluated using an organoid-lymphocyte coculture and a nucleotide-binding oligomerisation domain, leucine-rich repeat and CARD domain containing 5 (NLRC5) dextran sulphate sodium (DSS) colitis knock-out mouse model.
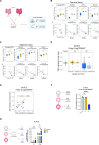
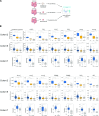
Results: We identified highly stable, CD-associated loss of DNAm at major histocompatibility complex (MHC) class 1 loci including NLRC5 and cognate gene upregulation. Single-cell RNA sequencing of primary mucosal tissue and IEOs confirmed the role of NLRC5 as transcriptional transactivator in the intestinal epithelium. Increased mucosal MHC-I and NLRC5 expression in adult and paediatric patients with CD was validated in additional cohorts and the functional role of MHC-I highlighted by demonstrating a relative protection from DSS-mediated mucosal inflammation in NLRC5-deficient mice. MHC-I DNAm in IEOs showed a significant correlation with CD disease phenotype and outcomes. Application of machine learning approaches enabled the development of a disease prognostic epigenetic molecular signature.
Conclusions: Our study has identified epigenetically regulated intestinal epithelial MHC-I as a novel mechanism in CD pathogenesis.
Keywords: CROHN'S DISEASE; INFLAMMATORY BOWEL DISEASE; INTESTINAL EPITHELIUM; METHYLATION.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures





References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
