The nucleolar phase of signal recognition particle assembly
- PMID: 38858088
- PMCID: PMC11165425
- DOI: 10.26508/lsa.202402614
The nucleolar phase of signal recognition particle assembly
Abstract
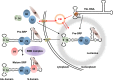
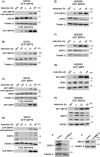
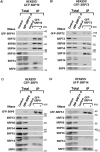
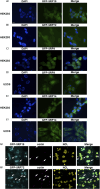
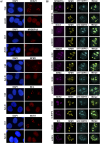
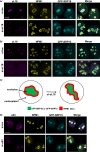
The signal recognition particle is essential for targeting transmembrane and secreted proteins to the endoplasmic reticulum. Remarkably, because they work together in the cytoplasm, the SRP and ribosomes are assembled in the same biomolecular condensate: the nucleolus. How important is the nucleolus for SRP assembly is not known. Using quantitative proteomics, we have investigated the interactomes of SRP components. We reveal that SRP proteins are associated with scores of nucleolar proteins important for ribosome biogenesis and nucleolar structure. Having monitored the subcellular distribution of SRP proteins upon controlled nucleolar disruption, we conclude that an intact organelle is required for their proper localization. Lastly, we have detected two SRP proteins in Cajal bodies, which indicates that previously undocumented steps of SRP assembly may occur in these bodies. This work highlights the importance of a structurally and functionally intact nucleolus for efficient SRP production and suggests that the biogenesis of SRP and ribosomes may be coordinated in the nucleolus by common assembly factors.
© 2024 Issa et al.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures








Similar articles
-
Biogenesis of the signal recognition particle (SRP) involves import of SRP proteins into the nucleolus, assembly with the SRP-RNA, and Xpo1p-mediated export.J Cell Biol. 2001 May 14;153(4):745-62. doi: 10.1083/jcb.153.4.745. J Cell Biol. 2001. PMID: 11352936 Free PMC article.
-
Signal recognition particle components in the nucleolus.Proc Natl Acad Sci U S A. 2000 Jan 4;97(1):55-60. doi: 10.1073/pnas.97.1.55. Proc Natl Acad Sci U S A. 2000. PMID: 10618370 Free PMC article.
-
Signal recognition particle RNA localization within the nucleolus differs from the classical sites of ribosome synthesis.J Cell Biol. 2002 Nov 11;159(3):411-8. doi: 10.1083/jcb.200208037. Epub 2002 Nov 11. J Cell Biol. 2002. PMID: 12427865 Free PMC article.
-
In vivo assembly of eukaryotic signal recognition particle: A still enigmatic process involving the SMN complex.Biochimie. 2019 Sep;164:99-104. doi: 10.1016/j.biochi.2019.04.007. Epub 2019 Apr 9. Biochimie. 2019. PMID: 30978374 Review.
-
Conventional and nonconventional roles of the nucleolus.Int Rev Cytol. 2002;219:199-266. doi: 10.1016/s0074-7696(02)19014-0. Int Rev Cytol. 2002. PMID: 12211630 Free PMC article. Review.
Cited by
-
Differentially expressed ncRNAs as key regulators in infection of human bronchial epithelial cells by the SARS-CoV-2 Delta variant.Mol Ther Nucleic Acids. 2025 May 14;36(2):102559. doi: 10.1016/j.omtn.2025.102559. eCollection 2025 Jun 10. Mol Ther Nucleic Acids. 2025. PMID: 40510596 Free PMC article.
-
Signal Peptides: From Molecular Mechanisms to Applications in Protein and Vaccine Engineering.Biomolecules. 2025 Jun 18;15(6):897. doi: 10.3390/biom15060897. Biomolecules. 2025. PMID: 40563537 Free PMC article. Review.
References
-
- Boulon S, Pradet-Balade B, Verheggen C, Molle D, Boireau S, Georgieva M, Azzag K, Robert MC, Ahmad Y, Neel H, et al. (2010. a) HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol Cell 39: 912–924. 10.1016/j.molcel.2010.08.023 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
