Pharmacological modulation of developmental and synaptic phenotypes in human SHANK3 deficient stem cell-derived neuronal models
- PMID: 38858349
- PMCID: PMC11165012
- DOI: 10.1038/s41398-024-02947-3
Pharmacological modulation of developmental and synaptic phenotypes in human SHANK3 deficient stem cell-derived neuronal models
Abstract
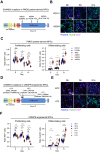
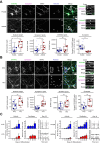
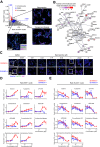
Phelan-McDermid syndrome (PMDS) arises from mutations in the terminal region of chromosome 22q13, impacting the SHANK3 gene. The resulting deficiency of the postsynaptic density scaffolding protein SHANK3 is associated with autism spectrum disorder (ASD). We examined 12 different PMDS patient and CRISPR-engineered stem cell-derived neuronal models and controls and found that reduced expression of SHANK3 leads to neuronal hyperdifferentiation, increased synapse formation, and decreased neuronal activity. We performed automated imaging-based screening of 7,120 target-annotated small molecules and identified three compounds that rescued SHANK3-dependent neuronal hyperdifferentiation. One compound, Benproperine, rescued the decreased colocalization of Actin Related Protein 2/3 Complex Subunit 2 (ARPC2) with ß-actin and rescued increased synapse formation in SHANK3 deficient neurons when administered early during differentiation. Neuronal activity was only mildly affected, highlighting Benproperine's effects as a neurodevelopmental modulator. This study demonstrates that small molecular compounds that reverse developmental phenotypes can be identified in human neuronal PMDS models.
© 2024. The Author(s).
Conflict of interest statement
AT, KS, DH, LCo, AW, AV, CJ, PS, and JW are or were employees of Ksilink. The remaining authors have no conflicts of interest to declare.
Figures



References
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

