Retrotransposons in Werner syndrome-derived macrophages trigger type I interferon-dependent inflammation in an atherosclerosis model
- PMID: 38858384
- PMCID: PMC11164933
- DOI: 10.1038/s41467-024-48663-w
Retrotransposons in Werner syndrome-derived macrophages trigger type I interferon-dependent inflammation in an atherosclerosis model
Abstract
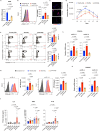
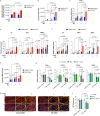
The underlying mechanisms of atherosclerosis, the second leading cause of death among Werner syndrome (WS) patients, are not fully understood. Here, we establish an in vitro co-culture system using macrophages (iMφs), vascular endothelial cells (iVECs), and vascular smooth muscle cells (iVSMCs) derived from induced pluripotent stem cells. In co-culture, WS-iMφs induces endothelial dysfunction in WS-iVECs and characteristics of the synthetic phenotype in WS-iVSMCs. Transcriptomics and open chromatin analysis reveal accelerated activation of type I interferon signaling and reduced chromatin accessibility of several transcriptional binding sites required for cellular homeostasis in WS-iMφs. Furthermore, the H3K9me3 levels show an inverse correlation with retrotransposable elements, and retrotransposable element-derived double-stranded RNA activates the DExH-box helicase 58 (DHX58)-dependent cytoplasmic RNA sensing pathway in WS-iMφs. Conversely, silencing type I interferon signaling in WS-iMφs rescues cell proliferation and suppresses cellular senescence and inflammation. These findings suggest that Mφ-specific inhibition of type I interferon signaling could be targeted to treat atherosclerosis in WS patients.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Goto M, Ishikawa Y, Sugimoto M, Furuichi Y. Werner syndrome: A changing pattern of clinical manifestations in Japan (1917\ 2008) Biosci. Trends. 2013;7:13–22. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials

