A refined picture of the native amine dehydrogenase family revealed by extensive biodiversity screening
- PMID: 38858403
- PMCID: PMC11164908
- DOI: 10.1038/s41467-024-49009-2
A refined picture of the native amine dehydrogenase family revealed by extensive biodiversity screening
Abstract
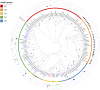
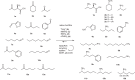
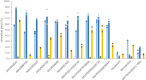
Native amine dehydrogenases offer sustainable access to chiral amines, so the search for scaffolds capable of converting more diverse carbonyl compounds is required to reach the full potential of this alternative to conventional synthetic reductive aminations. Here we report a multidisciplinary strategy combining bioinformatics, chemoinformatics and biocatalysis to extensively screen billions of sequences in silico and to efficiently find native amine dehydrogenases features using computational approaches. In this way, we achieve a comprehensive overview of the initial native amine dehydrogenase family, extending it from 2,011 to 17,959 sequences, and identify native amine dehydrogenases with non-reported substrate spectra, including hindered carbonyls and ethyl ketones, and accepting methylamine and cyclopropylamine as amine donor. We also present preliminary model-based structural information to inform the design of potential (R)-selective amine dehydrogenases, as native amine dehydrogenases are mostly (S)-selective. This integrated strategy paves the way for expanding the resource of other enzyme families and in highlighting enzymes with original features.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Development of an amine dehydrogenase for synthesis of chiral amines.Angew Chem Int Ed Engl. 2012 Apr 16;51(16):3969-72. doi: 10.1002/anie.201107813. Epub 2012 Mar 6. Angew Chem Int Ed Engl. 2012. PMID: 22396126
-
Incorporating Fast Protein Dynamics into Enzyme Design: A Proposed Mutant Aromatic Amine Dehydrogenase.J Phys Chem B. 2017 Aug 3;121(30):7290-7298. doi: 10.1021/acs.jpcb.7b05319. Epub 2017 Jul 19. J Phys Chem B. 2017. PMID: 28696108 Free PMC article.
-
Structure and Mutation of the Native Amine Dehydrogenase MATOUAmDH2.Chembiochem. 2022 May 18;23(10):e202200136. doi: 10.1002/cbic.202200136. Epub 2022 Apr 7. Chembiochem. 2022. PMID: 35349204 Free PMC article.
-
Recent achievements in developing the biocatalytic toolbox for chiral amine synthesis.Curr Opin Chem Biol. 2014 Apr;19:180-92. doi: 10.1016/j.cbpa.2014.02.021. Epub 2014 Apr 12. Curr Opin Chem Biol. 2014. PMID: 24721252 Review.
-
NAD(P)H-Dependent Dehydrogenases for the Asymmetric Reductive Amination of Ketones: Structure, Mechanism, Evolution and Application.Adv Synth Catal. 2017 Jun 19;359(12):2011-2025. doi: 10.1002/adsc.201700356. Epub 2017 May 11. Adv Synth Catal. 2017. PMID: 30008635 Free PMC article. Review.
Cited by
-
ASMC: investigating the amino acid diversity of enzyme active sites.Bioinformatics. 2025 Jun 2;41(6):btaf307. doi: 10.1093/bioinformatics/btaf307. Bioinformatics. 2025. PMID: 40372409 Free PMC article.
-
Selective hydrogenation of nitro compounds to amines by coupled redox reactions over a heterogeneous biocatalyst.Nat Commun. 2024 Aug 24;15(1):7297. doi: 10.1038/s41467-024-51531-2. Nat Commun. 2024. PMID: 39181899 Free PMC article.
-
An Update: Enzymatic Synthesis for Industrial Applications.Angew Chem Int Ed Engl. 2025 Jul;64(27):e202505976. doi: 10.1002/anie.202505976. Epub 2025 May 16. Angew Chem Int Ed Engl. 2025. PMID: 40241335 Free PMC article. Review.
-
In situ characterization of amine-forming enzymes shows altered oligomeric state.Protein Sci. 2025 Jan;34(1):e5248. doi: 10.1002/pro.5248. Protein Sci. 2025. PMID: 39720905 Free PMC article.
References
-
- Hughes DL. Highlights of the recent patent literature─focus on biocatalysis innovation. Org. Process Res. Dev. 2022;26:1878–1899. doi: 10.1021/acs.oprd.1c00417. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

