T-cell immunity induced and reshaped by an anti-HPV immuno-oncotherapeutic lentiviral vector
- PMID: 38858404
- PMCID: PMC11164992
- DOI: 10.1038/s41541-024-00894-0
T-cell immunity induced and reshaped by an anti-HPV immuno-oncotherapeutic lentiviral vector
Abstract
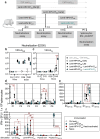
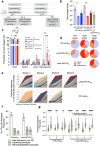
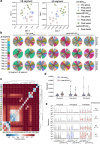
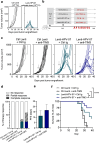
We recently developed an immuno-oncotherapy against human papillomavirus (HPV)-induced tumors based on a lentiviral vector encoding the Early E6 and E7 oncoproteins of HPV16 and HPV18 genotypes, namely "Lenti-HPV-07". The robust and long-lasting anti-tumor efficacy of Lenti-HPV-07 is dependent on CD8+ T-cell induction and remodeling of the tumor microenvironment. Here, we first established that anti-vector immunity induced by Lenti-HPV-07 prime has no impact on the efficacy of a homologous boost to amplify anti-HPV T-cell immunity. To longitudinally monitor the evolution of the T-cell repertoire generated after the prime, homologous or heterologous boost with Lenti-HPV-07, we tracked T-cell clonotypes by deep sequencing of T-Cell Receptor (TCR) variable β and α chain mRNA, applied to whole peripheral blood cells (PBL) and a T cell population specific of an immunodominant E7HPV16 epitope. We observed a hyper-expansion of clonotypes post prime, accompanied by increased frequencies of HPV-07-specific T cells. Additionally, there was a notable diversification of clonotypes post boost in whole PBL, but not in the E7HPV16-specific T cells. We then demonstrated that the effector functions of such Lenti-HPV-07-induced T cells synergize with anti-checkpoint inhibitory treatments by systemic administration of anti-TIM3 or anti-NKG2A monoclonal antibodies. While Lenti-HPV-07 is about to enter a Phase I/IIa clinical trial, these results will help better elucidate its mode of action in immunotherapy against established HPV-mediated malignancies.
© 2024. The Author(s).
Conflict of interest statement
P.C. is the founder and CSO of TheraVectys. I.F., L.D., B.V., F.M., A.N., P.A., F.L.C. and F.A. are employees of TheraVectys. L.M. has a consultancy activity for TheraVectys. I.F., L.D., F.M., A.N., P.C., L.M. and F.A. are inventors of a pending patent directed to the potential of Lenti-HPV-07 vaccination against HPV-induced cancers. Other authors declare no competing interests.
Figures

Similar articles
-
Full eradication of pre-clinical human papilloma virus-induced tumors by a lentiviral vaccine.EMBO Mol Med. 2023 Oct 11;15(10):e17723. doi: 10.15252/emmm.202317723. Epub 2023 Sep 7. EMBO Mol Med. 2023. PMID: 37675835 Free PMC article.
-
Optimization of heterologous DNA-prime, protein boost regimens and site of vaccination to enhance therapeutic immunity against human papillomavirus-associated disease.Cell Biosci. 2016 Feb 25;6:16. doi: 10.1186/s13578-016-0080-z. eCollection 2016. Cell Biosci. 2016. PMID: 26918115 Free PMC article.
-
CD4+ T-cell epitope-based heterologous prime-boost vaccination potentiates anti-tumor immunity and PD-1/PD-L1 immunotherapy.J Immunother Cancer. 2022 May;10(5):e004022. doi: 10.1136/jitc-2021-004022. J Immunother Cancer. 2022. PMID: 35580929 Free PMC article.
-
TriVax-HPV: an improved peptide-based therapeutic vaccination strategy against human papillomavirus-induced cancers.Cancer Immunol Immunother. 2012 Aug;61(8):1307-17. doi: 10.1007/s00262-012-1259-8. Epub 2012 Apr 22. Cancer Immunol Immunother. 2012. PMID: 22527249 Free PMC article. Review.
-
Development and Characterization of a Novel Non-Lytic Cancer Immunotherapy Using a Recombinant Arenavirus Vector Platform.Front Oncol. 2021 Oct 14;11:732166. doi: 10.3389/fonc.2021.732166. eCollection 2021. Front Oncol. 2021. PMID: 34722273 Free PMC article. Review.
Cited by
-
A lentiviral vector targeting a KRAS neoepitope for cancer immunotherapy.Sci Rep. 2025 Jul 2;15(1):23171. doi: 10.1038/s41598-025-05134-6. Sci Rep. 2025. PMID: 40603894 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Research Materials

