Efficient site-specific integration of large genes in mammalian cells via continuously evolved recombinases and prime editing
- PMID: 38858586
- PMCID: PMC11754103
- DOI: 10.1038/s41551-024-01227-1
Efficient site-specific integration of large genes in mammalian cells via continuously evolved recombinases and prime editing
Abstract
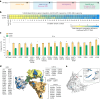
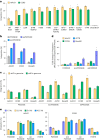
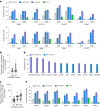
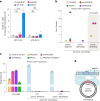
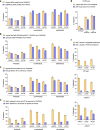
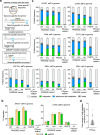
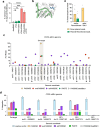
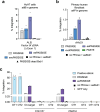
Methods for the targeted integration of genes in mammalian genomes suffer from low programmability, low efficiencies or low specificities. Here we show that phage-assisted continuous evolution enhances prime-editing-assisted site-specific integrase gene editing (PASSIGE), which couples the programmability of prime editing with the ability of recombinases to precisely integrate large DNA cargoes exceeding 10 kilobases. Evolved and engineered Bxb1 recombinase variants (evoBxb1 and eeBxb1) mediated up to 60% donor integration (3.2-fold that of wild-type Bxb1) in human cell lines with pre-installed recombinase landing sites. In single-transfection experiments at safe-harbour and therapeutically relevant sites, PASSIGE with eeBxb1 led to an average targeted-gene-integration efficiencies of 23% (4.2-fold that of wild-type Bxb1). Notably, integration efficiencies exceeded 30% at multiple sites in primary human fibroblasts. PASSIGE with evoBxb1 or eeBxb1 outperformed PASTE (for 'programmable addition via site-specific targeting elements', a method that uses prime editors fused to recombinases) on average by 9.1-fold and 16-fold, respectively. PASSIGE with continuously evolved recombinases is an unusually efficient method for the targeted integration of genes in mammalian cells.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: X.D.G., S.P. and D.R.L. have filed patent applications on this work. M.J.O. receives compensation as a consultant for Agathos Biologics. D.R.L. is a co-founder and consultant for Prime Medicine, Beam Therapeutics, Pairwise Plants, Chroma Medicine and Nvelop Therapeutics, and owns equity in these companies.
Figures



References
-
- Weischenfeldt, J., Symmons, O., Spitz, F. & Korbel, J. O. Phenotypic impact of genomic structural variation: insights from and for human disease. Nat. Rev. Genet.14, 125–138 (2013). - PubMed
-
- Bareil, C. & Bergougnoux, A. CFTR gene variants, epidemiology and molecular pathology. Arch. Pediatr.27, eS8–eS12 (2020). - PubMed
-
- Jin, X. et al. Identification of novel deep intronic PAH gene variants in patients diagnosed with phenylketonuria. Hum. Mutat.43, 56–66 (2022). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

