Functional and structural basis of human parainfluenza virus type 3 neutralization with human monoclonal antibodies
- PMID: 38858594
- PMCID: PMC12203573
- DOI: 10.1038/s41564-024-01722-w
Functional and structural basis of human parainfluenza virus type 3 neutralization with human monoclonal antibodies
Abstract
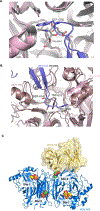
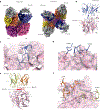
Human parainfluenza virus type 3 (hPIV3) is a respiratory pathogen that can cause severe disease in older people and infants. Currently, vaccines against hPIV3 are in clinical trials but none have been approved yet. The haemagglutinin-neuraminidase (HN) and fusion (F) surface glycoproteins of hPIV3 are major antigenic determinants. Here we describe naturally occurring potently neutralizing human antibodies directed against both surface glycoproteins of hPIV3. We isolated seven neutralizing HN-reactive antibodies and a pre-fusion conformation F-reactive antibody from human memory B cells. One HN-binding monoclonal antibody (mAb), designated PIV3-23, exhibited functional attributes including haemagglutination and neuraminidase inhibition. We also delineated the structural basis of neutralization for two HN and one F mAbs. MAbs that neutralized hPIV3 in vitro protected against infection and disease in vivo in a cotton rat model of hPIV3 infection, suggesting correlates of protection for hPIV3 and the potential clinical utility of these mAbs.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Conflict of interest statement
Competing interests
J.E.C. has served as a consultant for Luna Labs USA, Merck Sharp & Dohme Corporation, Emergent Biosolutions, and BTG International Inc., is a member of the Scientific Advisory Board of Meissa Vaccines, a former member of the Scientific Advisory Board of Gigagen (Grifols) and is founder of IDBiologics. The laboratory of J.E.C. received unrelated sponsored research agreements from AstraZeneca, Takeda Vaccines, and IDBiologics during the conduct of the study. T.S.J. has served as a consultant for Pfizer. A.L.G reports contract testing from Abbott, Cepheid, Novavax, Pfizer, Janssen and Hologic, and research support from Gilead, outside of the described work. A.M. reports potential future financial interests in Thylacine Biotherapeutics Inc. unrelated to the present study. All other authors declare no competing interests. Vanderbilt University has applied for a patent for some of the antibodies in this paper (2024 International Patent Application No. PCT/US2024/028239 based on US Serial No. 63/504,549).
Figures





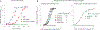




References
-
- Chanock RM et al. Newly recognized myxoviruses from children with respiratory disease. N. Engl. J. Med 258, 207–213 (1958). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

