ASS1 metabolically contributes to the nuclear and cytosolic p53-mediated DNA damage response
- PMID: 38858597
- PMCID: PMC11272581
- DOI: 10.1038/s42255-024-01060-5
ASS1 metabolically contributes to the nuclear and cytosolic p53-mediated DNA damage response
Erratum in
-
Author Correction: ASS1 metabolically contributes to the nuclear and cytosolic p53-mediated DNA damage response.Nat Metab. 2024 Jul;6(7):1417. doi: 10.1038/s42255-024-01090-z. Nat Metab. 2024. PMID: 38965441 Free PMC article. No abstract available.
Abstract
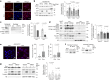
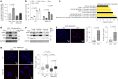
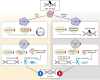
Downregulation of the urea cycle enzyme argininosuccinate synthase (ASS1) in multiple tumors is associated with a poor prognosis partly because of the metabolic diversion of cytosolic aspartate for pyrimidine synthesis, supporting proliferation and mutagenesis owing to nucleotide imbalance. Here, we find that prolonged loss of ASS1 promotes DNA damage in colon cancer cells and fibroblasts from subjects with citrullinemia type I. Following acute induction of DNA damage with doxorubicin, ASS1 expression is elevated in the cytosol and the nucleus with at least a partial dependency on p53; ASS1 metabolically restrains cell cycle progression in the cytosol by restricting nucleotide synthesis. In the nucleus, ASS1 and ASL generate fumarate for the succination of SMARCC1, destabilizing the chromatin-remodeling complex SMARCC1-SNF5 to decrease gene transcription, specifically in a subset of the p53-regulated cell cycle genes. Thus, following DNA damage, ASS1 is part of the p53 network that pauses cell cycle progression, enabling genome maintenance and survival. Loss of ASS1 contributes to DNA damage and promotes cell cycle progression, likely contributing to cancer mutagenesis and, hence, adaptability potential.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis.Nature. 2015 Nov 19;527(7578):379-383. doi: 10.1038/nature15529. Epub 2015 Nov 11. Nature. 2015. PMID: 26560030 Free PMC article.
-
Argininosuccinate synthase 1 is an intrinsic Akt repressor transactivated by p53.Sci Adv. 2017 May 19;3(5):e1603204. doi: 10.1126/sciadv.1603204. eCollection 2017 May. Sci Adv. 2017. PMID: 28560349 Free PMC article.
-
ASS1 and ASL suppress growth in clear cell renal cell carcinoma via altered nitrogen metabolism.Cancer Metab. 2021 Dec 3;9(1):40. doi: 10.1186/s40170-021-00271-8. Cancer Metab. 2021. PMID: 34861885 Free PMC article.
-
Advances in the impact of ASS1 dysregulation on metabolic reprogramming of tumor cells.Cell Signal. 2025 Mar;127:111593. doi: 10.1016/j.cellsig.2025.111593. Epub 2025 Jan 6. Cell Signal. 2025. PMID: 39778698 Review.
-
Mutations and polymorphisms in the human argininosuccinate synthetase (ASS1) gene.Hum Mutat. 2009 Mar;30(3):300-7. doi: 10.1002/humu.20847. Hum Mutat. 2009. PMID: 19006241 Review.
Cited by
-
Etoposide-induced protein 2.4 homolog promotes argininosuccinate synthase 1 and cancer cell survival upon arginine deprivation.Cell Mol Biol Lett. 2025 Apr 19;30(1):52. doi: 10.1186/s11658-025-00726-6. Cell Mol Biol Lett. 2025. PMID: 40253325 Free PMC article.
-
Metabolomic analysis reveals key changes in amino acid metabolism in colorectal cancer patients.Amino Acids. 2025 May 2;57(1):22. doi: 10.1007/s00726-025-03448-3. Amino Acids. 2025. PMID: 40314699 Free PMC article.
-
Arginine: at the crossroads of nitrogen metabolism.EMBO J. 2025 Mar;44(5):1275-1293. doi: 10.1038/s44318-025-00379-3. Epub 2025 Feb 7. EMBO J. 2025. PMID: 39920310 Free PMC article. Review.
-
Dynamic crosstalk between amino acid metabolism and cancer drug efficacy: From mechanisms to therapeutic opportunities.iScience. 2025 Apr 11;28(5):112405. doi: 10.1016/j.isci.2025.112405. eCollection 2025 May 16. iScience. 2025. PMID: 40625405 Free PMC article. Review.
-
ASS1 is a hub gene and possible therapeutic target for regulating metabolic dysfunction-associated steatotic liver disease modulated by a carbohydrate-restricted diet.Mol Divers. 2025 Aug;29(4):3717-3732. doi: 10.1007/s11030-025-11187-6. Epub 2025 Apr 17. Mol Divers. 2025. PMID: 40244373
References
-
- Ah Mew, N et al. Urea cycle disorders overview in GeneReviews (eds. Adam M. P. et al.) (University of Washington, 2017).
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

