CDK7/CDK9 mediates transcriptional activation to prime paraptosis in cancer cells
- PMID: 38858714
- PMCID: PMC11163730
- DOI: 10.1186/s13578-024-01260-2
CDK7/CDK9 mediates transcriptional activation to prime paraptosis in cancer cells
Abstract
Background: Paraptosis is a programmed cell death characterized by cytoplasmic vacuolation, which has been explored as an alternative method for cancer treatment and is associated with cancer resistance. However, the mechanisms underlying the progression of paraptosis in cancer cells remain largely unknown.
Methods: Paraptosis-inducing agents, CPYPP, cyclosporin A, and curcumin, were utilized to investigate the underlying mechanism of paraptosis. Next-generation sequencing and liquid chromatography-mass spectrometry analysis revealed significant changes in gene and protein expressions. Pharmacological and genetic approaches were employed to elucidate the transcriptional events related to paraptosis. Xenograft mouse models were employed to evaluate the potential of paraptosis as an anti-cancer strategy.
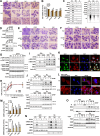
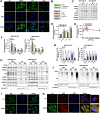
Results: CPYPP, cyclosporin A, and curcumin induced cytoplasmic vacuolization and triggered paraptosis in cancer cells. The paraptotic program involved reactive oxygen species (ROS) provocation and the activation of proteostatic dynamics, leading to transcriptional activation associated with redox homeostasis and proteostasis. Both pharmacological and genetic approaches suggested that cyclin-dependent kinase (CDK) 7/9 drive paraptotic progression in a mutually-dependent manner with heat shock proteins (HSPs). Proteostatic stress, such as accumulated cysteine-thiols, HSPs, ubiquitin-proteasome system, endoplasmic reticulum stress, and unfolded protein response, as well as ROS provocation primarily within the nucleus, enforced CDK7/CDK9-Rpb1 (RNAPII subunit B1) activation by potentiating its interaction with HSPs and protein kinase R in a forward loop, amplifying transcriptional regulation and thereby exacerbating proteotoxicity leading to initiate paraptosis. The xenograft mouse models of MDA-MB-231 breast cancer and docetaxel-resistant OECM-1 head and neck cancer cells further confirmed the induction of paraptosis against tumor growth.
Conclusions: We propose a novel regulatory paradigm in which the activation of CDK7/CDK9-Rpb1 by nuclear proteostatic stress mediates transcriptional regulation to prime cancer cell paraptosis.
Keywords: Cancer; Cyclin-dependent kinase 7/9; Heat shock proteins; Nuclear stress; Paraptosis; Protein kinase R; Reactive oxygen species.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures







References
-
- Wang Y, Li X, Wang L, Ding P, Zhang Y, Han W, et al. An alternative form of paraptosis-like cell death, triggered by TAJ/TROY and enhanced by PDCD5 overexpression. J Cell Sci. 2004;117(8):1525–32. - PubMed
Grants and funding
- MOST 109-2320-B-039-016/Ministry of Science and Technology, Taiwan
- MOST 110-2320-B-039-013-MY3/Ministry of Science and Technology, Taiwan
- NSTC 112-2313-B-005-039/Ministry of Science and Technology, Taiwan
- Cancer Biology and Precision Therapeutics Center/Ministry of Education, Taiwan
- MOE-112-S-0023-A/Ministry of Education, Taiwan
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

