Coral microbiomes are structured by environmental gradients in deep waters
- PMID: 38858739
- PMCID: PMC11165896
- DOI: 10.1186/s40793-024-00579-0
Coral microbiomes are structured by environmental gradients in deep waters
Abstract
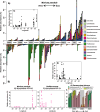
Background: Coral-associated microbiomes vary greatly between colonies and localities with functional consequences on the host. However, the full extent of variability across the ranges of most coral species remains unknown, especially for corals living in deep waters which span greater ranges. Here, we characterized the microbiomes of four octocoral species from mesophotic and bathyal deep-sea habitats in the northern Gulf of Mexico, Muricea pendula, Swiftia exserta, Callogorgia delta, and Paramuricea biscaya, using 16S rRNA gene metabarcoding. We sampled extensively across their ranges to test for microbiome differentiation between and within species, examining the influence of environmental factors that vary with depth (53-2224 m) and geographic location (over 680 m) as well as the host coral's genotype using RAD-sequencing.
Results: Coral microbiomes were often dominated by amplicon sequence variants whose abundances varied across their hosts' ranges, including symbiotic taxa: corallicolids, Endozoicomonas, members of the Mollicutes, and the BD1-7 clade. Coral species, depth, and geographic location significantly affected diversity, microbial community composition, and the relative abundance of individual microbes. Depth was the strongest environmental factor determining microbiome structure within species, which influenced the abundance of most dominant symbiotic taxa. Differences in host genotype, bottom temperature, and surface primary productivity could explain a significant part of the microbiome variation associated with depth and geographic location.
Conclusions: Altogether, this work demonstrates that the microbiomes of corals in deep waters vary substantially across their ranges in accordance with depth and other environmental conditions. It reveals that the influence of depth on the ecology of mesophotic and deep-sea corals extends to its effects on their microbiomes which may have functional consequences. This work also identifies the distributions of microbes including potential parasites which can be used to inform restoration plans in response to the Deepwater Horizon oil spill.
Keywords: Deep-sea; Mesophotic; Microbial biogeography; Mycoplasma; Oceanoplasma; Seascape ecology.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Wegley L, Yu Y, Breitbart M, Casas V, Kline DI, Rohwer F. Coral-associated archaea. Mar Ecol Prog Ser. 2004;273:89–96. doi: 10.3354/meps273089. - DOI
Grants and funding
LinkOut - more resources
Full Text Sources
