Establishment of a circRNA-regulated E3 ubiquitin ligase signature and nomogram to predict immunotherapeutic efficacy and prognosis in hepatocellular carcinoma
- PMID: 38858746
- PMCID: PMC11163726
- DOI: 10.1186/s40001-024-01893-6
Establishment of a circRNA-regulated E3 ubiquitin ligase signature and nomogram to predict immunotherapeutic efficacy and prognosis in hepatocellular carcinoma
Abstract
Background: Hepatocellular carcinoma (HCC) is a common type of malignant tumor where the prognosis is dismal. Circular RNA (CircRNA) is a novel RNA that regulates downstream gene transcription and translation to influence the progression of HCC. However, the regulatory relationship that exists between E3 ligases, which is a class of post-translational modifying proteins, and circRNA remains unclear.
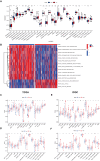
Methods: Based on the E3 ubiquitin ligase in the competitive endogenous RNA (ceRNA) network, a circRNA-regulated E3 ubiquitin ligase signature (CRE3UL) was developed. A CRE3UL signature was created using the least absolute shrinkage and selection operator (Lasso) and Cox regression analysis and merged it with clinicopathologic characteristics to generate a nomogram for prognosis prediction. The pRRophetic algorithm was utilized and immunological checkpoints were analyzed to compare the responses of patients in the high-risk group (HRG) and low-risk group (LRG) to targeted therapy and immunotherapy. Finally, experimental research will further elucidate the relationship between E3 ubiquitin ligase signature and HCC.
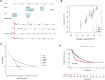
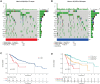
Results: HRG patients were found to have a worse prognosis than LRG patients. Furthermore, significant variations in prognosis were observed among different subgroups based on various clinical characteristics. The CRE3UL signature was identified as being an independent prognostic indicator. The nomogram that combined clinical characteristics and the CRE3UL signature was found to accurately predict the prognosis of HCC patients and demonstrated greater clinical utility than the current TNM staging approach. According to anticancer medication sensitivity predictions, the tumors of HRG patients were more responsive to gefitinib and nilotinib. From immune-checkpoint markers analysis, immunotherapy was identified as being more probable to assist those in the HRG.
Conclusions: We found a significant correlation between the CRE3UL signature and the tumor microenvironment, enabling precise prognosis prediction for HCC patients. Additionally, a nomogram was developed that performs well in predicting the overall survival (OS) of HCC patients. This provides valuable guidance for clinicians in devising specific personalized treatment strategies.
Keywords: CircRNA; E3 ubiquitin ligase signature; Hepatocellular carcinoma; Immune and prognosis; Nomogram.
© 2024. The Author(s).
Conflict of interest statement
The authors have no competing of interests or financial ties to disclose.
Figures



References
-
- Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Nikšić M, Bonaventure A, Valkov M, Johnson CJ, Estève J, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. The Lancet. 2018;391:1023–1075. doi: 10.1016/S0140-6736(17)33326-3. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

