Genome-wide DNA methylation in relation to ARID1A deficiency in ovarian clear cell carcinoma
- PMID: 38858765
- PMCID: PMC11163774
- DOI: 10.1186/s12967-024-05311-7
Genome-wide DNA methylation in relation to ARID1A deficiency in ovarian clear cell carcinoma
Abstract
Background: The poor chemo-response and high DNA methylation of ovarian clear cell carcinoma (OCCC) have attracted extensive attentions. Recently, we revealed the mutational landscape of the human kinome and additional cancer-related genes and found deleterious mutations in ARID1A, a component of the SWI/SNF chromatin-remodeling complex, in 46% of OCCC patients. The present study aims to comprehensively investigate whether ARID1A loss and genome-wide DNA methylation are co-regulated in OCCC and identify putative therapeutic targets epigenetically regulated by ARID1A.
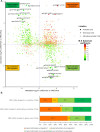
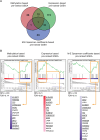
Methods: DNA methylation of ARID1Amt/ko and ARID1Awt OCCC tumors and cell lines were analyzed by Infinium MethylationEPIC BeadChip. The clustering of OCCC tumors in relation to clinical and mutational status of tumors were analyzed by hierarchical clustering analysis of genome-wide methylation. GEO expression profiles were used to identify differentially methylated (DM) genes and their expression level in ARID1Amt/ko vs ARID1Awt OCCCs. Combining three pre-ranked GSEAs, pathways and leading-edge genes epigenetically regulated by ARID1A were revealed. The leading-edge genes that passed the in-silico validation and showed consistent ARID1A-related methylation change in tumors and cell lines were regarded as candidate genes and finally verified by bisulfite sequencing and RT-qPCR.
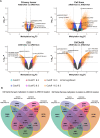
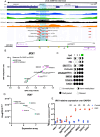
Results: Hierarchical clustering analysis of genome-wide methylation showed two clusters of OCCC tumors. Tumor stage, ARID1A/PIK3CA mutations and TP53 mutations were significantly different between the two clusters. ARID1A mutations in OCCC did not cause global DNA methylation changes but were related to DM promoter or gene-body CpG islands of 2004 genes. Three pre-ranked GSEAs collectively revealed the significant enrichment of EZH2- and H3K27me3-related gene-sets by the ARID1A-related DM genes. 13 Leading-edge DM genes extracted from the enriched gene-sets passed the expression-based in-silico validation and showed consistent ARID1A-related methylation change in tumors and cell lines. Bisulfite sequencing and RT-qPCR analysis showed promoter hypermethylation and lower expression of IRX1, TMEM101 and TRIP6 in ARID1Amt compared to ARID1Awt OCCC cells, which was reversed by 5-aza-2'-deoxycytidine treatment.
Conclusions: Our study shows that ARID1A loss is related to the differential methylation of a number of genes in OCCC. ARID1A-dependent DM genes have been identified as key genes of many cancer-related pathways that may provide new candidates for OCCC targeted treatment.
Keywords: ARID1A; DNA methylation; EZH2; Ovarian clear cell carcinoma.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures


Similar articles
-
Establishment and characterization of ovarian clear cell carcinoma patient-derived xenografts.Sci Rep. 2025 Feb 5;15(1):4434. doi: 10.1038/s41598-025-86384-2. Sci Rep. 2025. PMID: 39910133 Free PMC article.
-
DNA Methylation Profiles of Ovarian Clear Cell Carcinoma.Cancer Epidemiol Biomarkers Prev. 2022 Jan;31(1):132-141. doi: 10.1158/1055-9965.EPI-21-0677. Epub 2021 Oct 25. Cancer Epidemiol Biomarkers Prev. 2022. PMID: 34697060 Free PMC article.
-
Genomic profiling of ovarian clear cell carcinoma in Chinese patients reveals potential prognostic biomarkers for survival.Ann Med. 2023 Dec;55(1):2218104. doi: 10.1080/07853890.2023.2218104. Ann Med. 2023. PMID: 37272300 Free PMC article.
-
Precision medicine for ovarian clear cell carcinoma based on gene alterations.Int J Clin Oncol. 2020 Mar;25(3):419-424. doi: 10.1007/s10147-020-01622-z. Epub 2020 Feb 4. Int J Clin Oncol. 2020. PMID: 32020380 Review.
-
Potential therapeutic targets in ARID1A-mutated cancers.Expert Opin Ther Targets. 2015;19(11):1419-22. doi: 10.1517/14728222.2015.1062879. Epub 2015 Jun 30. Expert Opin Ther Targets. 2015. PMID: 26125128 Free PMC article. Review.
Cited by
-
Advances in research on malignant transformation of endometriosis-associated ovarian cancer.Front Oncol. 2024 Oct 9;14:1475231. doi: 10.3389/fonc.2024.1475231. eCollection 2024. Front Oncol. 2024. PMID: 39445058 Free PMC article. Review.
-
Establishment and characterization of ovarian clear cell carcinoma patient-derived xenografts.Sci Rep. 2025 Feb 5;15(1):4434. doi: 10.1038/s41598-025-86384-2. Sci Rep. 2025. PMID: 39910133 Free PMC article.
References
-
- Goff BA, Sainz De La Cuesta R, Muntz HG, Fleischhacker D, Ek M, Rice LW, et al. Clinical characteristics of clear cell carcinoma of the ovary: a distinct histologic type with poor prognosis and resistance to platinum-based chemotherapy. Gynecol Oncol. 1996;88(3):412–7. doi: 10.1006/gyno.1996.0065. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

