Coarse-grained modeling of annexin A2-induced microdomain formation on a vesicle
- PMID: 38859585
- PMCID: PMC11365106
- DOI: 10.1016/j.bpj.2024.06.006
Coarse-grained modeling of annexin A2-induced microdomain formation on a vesicle
Abstract
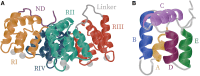
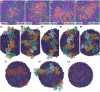
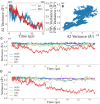
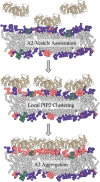
Annexin A2 (A2)-induced microdomain formation is a key step in biological processes such as Ca2+-mediated exocytosis in neuroendocrine cells. In this work, a total of 15 coarse-grained molecular dynamics simulations were performed on vesicle models having a diameter of approximately 250 Å for 15 μs each using the Martini2 force field. Five simulations were performed in the presence of 10 A2, 5 in the presence of A2 but absence of PIP2, and 5 simulations in the absence of A2 but presence of PIP2. Consistent results were generated among the simulations. A2-induced PIP2 microdomain formation was observed and shown to occur in three phases: A2-vesicle association, localized A2-induced PIP2 clustering, and A2 aggregation driving PIP2 microdomain formation. The relationship between A2 aggregation and PIP2 microdomain formation was quantitatively described using a novel method which calculated the variance among protein and lipid positions via the Fréchet mean. A large reduction in PIP2 variance was observed in the presence of A2 but not in its absence. This reduction in PIP2 variance was proportional to the reduction observed in A2 variance and demonstrates that the observed PIP2 microdomain formation is dependent upon A2 aggregation. The three-phase model of A2-induced microdomain formation generated in this work will serve as a valuable guide for further experimental studies and the development of novel A2 inhibitors. No microdomain formation was observed in the absence of A2 and minimal A2-membrane interaction was observed in the absence of PIP2.
Copyright © 2024 Biophysical Society. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures

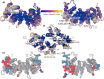
Similar articles
-
Lipid segregation and membrane budding induced by the peripheral membrane binding protein annexin A2.J Biol Chem. 2013 Aug 23;288(34):24764-76. doi: 10.1074/jbc.M113.474023. Epub 2013 Jul 16. J Biol Chem. 2013. PMID: 23861394 Free PMC article.
-
Membrane protein sequestering by ionic protein-lipid interactions.Nature. 2011 Oct 23;479(7374):552-5. doi: 10.1038/nature10545. Nature. 2011. PMID: 22020284 Free PMC article.
-
Membrane targeting of the yeast exocyst complex.Biochim Biophys Acta. 2015 Jul;1848(7):1481-9. doi: 10.1016/j.bbamem.2015.03.026. Epub 2015 Mar 30. Biochim Biophys Acta. 2015. PMID: 25838123
-
Annexin A2 extracellular translocation and virus interaction: A potential target for antivirus-drug discovery.Rev Med Virol. 2019 May;29(3):e2038. doi: 10.1002/rmv.2038. Epub 2019 Feb 11. Rev Med Virol. 2019. PMID: 30746844 Review.
-
Role of calcium in membrane interactions by PI(4,5)P₂-binding proteins.Biochem Soc Trans. 2014 Oct;42(5):1441-6. doi: 10.1042/BST20140149. Biochem Soc Trans. 2014. PMID: 25233429 Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

