Simultaneous Native Mass Spectrometry Analysis of Single and Double Mutants To Probe Lipid Binding to Membrane Proteins
- PMID: 38859611
- PMCID: PMC11215972
- DOI: 10.1021/acs.analchem.4c01704
Simultaneous Native Mass Spectrometry Analysis of Single and Double Mutants To Probe Lipid Binding to Membrane Proteins
Abstract
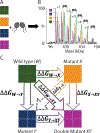
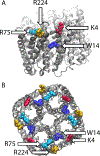
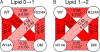
Lipids are critical modulators of membrane protein structure and function. However, it is challenging to investigate the thermodynamics of protein-lipid interactions because lipids can simultaneously bind membrane proteins at different sites with different specificities. Here, we developed a native mass spectrometry (MS) approach using single and double mutants to measure the relative energetic contributions of specific residues on Aquaporin Z (AqpZ) toward cardiolipin (CL) binding. We first mutated potential lipid-binding residues on AqpZ, and mixed mutant and wild-type proteins together with CL. By using native MS to simultaneously resolve lipid binding to the mutant and wild-type proteins in a single spectrum, we directly determined the relative affinities of CL binding, thereby revealing the relative Gibbs free energy change for lipid binding caused by the mutation. Comparing different mutants revealed that W14 contributes to the tightest CL binding site, with R224 contributing to a lower affinity site. Using double mutant cycling, we investigated the synergy between W14 and R224 sites on CL binding. Overall, this novel native MS approach provides unique insights into the binding of lipids to specific sites on membrane proteins.
Conflict of interest statement
Conflict of Interest
The authors declare no competing financial interest.
Figures

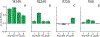
Update of
-
SIMULTANEOUS NATIVE MASS SPECTROMETRY ANALYSIS OF SINGLE AND DOUBLE MUTANTS TO PROBE LIPID BINDING TO MEMBRANE PROTEINS.bioRxiv [Preprint]. 2023 Dec 12:2023.09.19.558516. doi: 10.1101/2023.09.19.558516. bioRxiv. 2023. Update in: Anal Chem. 2024 Jun 25;96(25):10426-10433. doi: 10.1021/acs.analchem.4c01704. PMID: 37781586 Free PMC article. Updated. Preprint.
Similar articles
-
Alanine Scanning to Define Membrane Protein-Lipid Interaction Sites Using Native Mass Spectrometry.Biochemistry. 2025 Mar 18;64(6):1308-1316. doi: 10.1021/acs.biochem.4c00717. Epub 2025 Mar 6. Biochemistry. 2025. PMID: 40047061
-
SIMULTANEOUS NATIVE MASS SPECTROMETRY ANALYSIS OF SINGLE AND DOUBLE MUTANTS TO PROBE LIPID BINDING TO MEMBRANE PROTEINS.bioRxiv [Preprint]. 2023 Dec 12:2023.09.19.558516. doi: 10.1101/2023.09.19.558516. bioRxiv. 2023. Update in: Anal Chem. 2024 Jun 25;96(25):10426-10433. doi: 10.1021/acs.analchem.4c01704. PMID: 37781586 Free PMC article. Updated. Preprint.
-
Diagnostic test accuracy and cost-effectiveness of tests for codeletion of chromosomal arms 1p and 19q in people with glioma.Cochrane Database Syst Rev. 2022 Mar 2;3(3):CD013387. doi: 10.1002/14651858.CD013387.pub2. Cochrane Database Syst Rev. 2022. PMID: 35233774 Free PMC article.
-
Rufinamide add-on therapy for refractory epilepsy.Cochrane Database Syst Rev. 2018 Apr 25;4(4):CD011772. doi: 10.1002/14651858.CD011772.pub2. Cochrane Database Syst Rev. 2018. Update in: Cochrane Database Syst Rev. 2020 Nov 8;11:CD011772. doi: 10.1002/14651858.CD011772.pub3. PMID: 29691835 Free PMC article. Updated.
-
Drugs for preventing postoperative nausea and vomiting in adults after general anaesthesia: a network meta-analysis.Cochrane Database Syst Rev. 2020 Oct 19;10(10):CD012859. doi: 10.1002/14651858.CD012859.pub2. Cochrane Database Syst Rev. 2020. PMID: 33075160 Free PMC article.
Cited by
-
Double and triple thermodynamic mutant cycles reveal the basis for specific MsbA-lipid interactions.bioRxiv [Preprint]. 2023 Nov 6:2023.07.03.547565. doi: 10.1101/2023.07.03.547565. bioRxiv. 2023. Update in: Elife. 2024 Jan 22;12:RP91094. doi: 10.7554/eLife.91094. PMID: 37461710 Free PMC article. Updated. Preprint.
-
Alanine Scanning to Define Membrane Protein-Lipid Interaction Sites Using Native Mass Spectrometry.bioRxiv [Preprint]. 2025 Feb 3:2024.10.24.620105. doi: 10.1101/2024.10.24.620105. bioRxiv. 2025. Update in: Biochemistry. 2025 Mar 18;64(6):1308-1316. doi: 10.1021/acs.biochem.4c00717. PMID: 39484449 Free PMC article. Updated. Preprint.
-
Alanine Scanning to Define Membrane Protein-Lipid Interaction Sites Using Native Mass Spectrometry.Biochemistry. 2025 Mar 18;64(6):1308-1316. doi: 10.1021/acs.biochem.4c00717. Epub 2025 Mar 6. Biochemistry. 2025. PMID: 40047061
-
Grafting the ALFA tag for structural studies of aquaporin Z.J Struct Biol X. 2024 Feb 2;9:100097. doi: 10.1016/j.yjsbx.2024.100097. eCollection 2024 Jun. J Struct Biol X. 2024. PMID: 38361954 Free PMC article.
-
Native Top-Down Analysis of Membrane Protein Complexes Directly From In Vitro and Native Membranes.Mol Cell Proteomics. 2025 Jul;24(7):100993. doi: 10.1016/j.mcpro.2025.100993. Epub 2025 May 14. Mol Cell Proteomics. 2025. PMID: 40378922 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

