YajC, a predicted membrane protein, promotes Enterococcus faecium biofilm formation in vitro and in a rat endocarditis model
- PMID: 38860142
- PMCID: PMC11163983
- DOI: 10.1093/femsmc/xtae017
YajC, a predicted membrane protein, promotes Enterococcus faecium biofilm formation in vitro and in a rat endocarditis model
Abstract
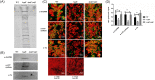
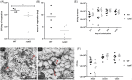
Biofilm formation is a critical step in the pathogenesis of difficult-to-treat Gram-positive bacterial infections. We identified that YajC, a conserved membrane protein in bacteria, plays a role in biofilm formation of the clinically relevant Enterococcus faecium strain E1162. Deletion of yajC conferred significantly impaired biofilm formation in vitro and was attenuated in a rat endocarditis model. Mass spectrometry analysis of supernatants of washed ΔyajC cells revealed increased amounts in cytoplasmic and cell-surface-located proteins, including biofilm-associated proteins, suggesting that proteins on the surface of the yajC mutant are only loosely attached. In Streptococcus mutans YajC has been identified in complex with proteins of two cotranslational membrane protein-insertion pathways; the signal recognition particle (SRP)-SecYEG-YajC-YidC1 and the SRP-YajC-YidC2 pathway, but its function is unknown. In S. mutans mutation of yidC1 and yidC2 resulted in impaired protein insertion in the cell membrane and secretion in the supernatant. The E. faecium genome contains all homologous genes encoding for the cotranslational membrane protein-insertion pathways. By combining the studies in S. mutans and E. faecium, we propose that YajC is involved in the stabilization of the SRP-SecYEG-YajC-YidC1 and SRP-YajC-Yid2 pathway or plays a role in retaining proteins for proper docking to the YidC insertases for translocation in and over the membrane.
Keywords: Biofilm; Enterococcus faecium; Streptococcus mutans; YajC; cotranslational membrane protein insertion pathway; rat endocarditis model.
© The Author(s) 2024. Published by Oxford University Press on behalf of FEMS.
Conflict of interest statement
The authors declare noncompeting interests.
Figures


Similar articles
-
Protein Interactomes of Streptococcus mutans YidC1 and YidC2 Membrane Protein Insertases Suggest SRP Pathway-Independent- and -Dependent Functions, Respectively.mSphere. 2021 Mar 3;6(2):e01308-20. doi: 10.1128/mSphere.01308-20. mSphere. 2021. PMID: 33658280 Free PMC article.
-
Membrane proteomic analysis reveals overlapping and independent functions of Streptococcus mutans Ffh, YidC1, and YidC2.Mol Oral Microbiol. 2019 Aug;34(4):131-152. doi: 10.1111/omi.12261. Epub 2019 Jun 7. Mol Oral Microbiol. 2019. PMID: 31034136 Free PMC article.
-
Streptococcus mutans yidC1 and yidC2 Impact Cell Envelope Biogenesis, the Biofilm Matrix, and Biofilm Biophysical Properties.J Bacteriol. 2018 Dec 7;201(1):e00396-18. doi: 10.1128/JB.00396-18. Print 2019 Jan 1. J Bacteriol. 2018. PMID: 30322852 Free PMC article.
-
Protein traffic in bacteria: multiple routes from the ribosome to and across the membrane.Prog Nucleic Acid Res Mol Biol. 2001;66:107-57. doi: 10.1016/s0079-6603(00)66028-2. Prog Nucleic Acid Res Mol Biol. 2001. PMID: 11051763 Review.
-
The Sec translocase.Biochim Biophys Acta. 2011 Mar;1808(3):851-65. doi: 10.1016/j.bbamem.2010.08.016. Epub 2010 Aug 27. Biochim Biophys Acta. 2011. PMID: 20801097 Review.
References
-
- Abee T, Kovács ÁT, Kuipers OP et al. Biofilm formation and dispersal in Gram-positive bacteria. Curr Opin Biotechnol. 2011;22:172–9. - PubMed
-
- Chmielewski RAN, Frank JF. Biofilm formation and control in food processing facilities. Compr Rev Food Sci Food Saf. 2003;2:22–32. - PubMed
-
- Costerton JW. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 2001;9:50–2. - PubMed
LinkOut - more resources
Full Text Sources
