The interferon-rich skin environment regulates Langerhans cell ADAM17 to promote photosensitivity in lupus
- PMID: 38860651
- PMCID: PMC11213570
- DOI: 10.7554/eLife.85914
The interferon-rich skin environment regulates Langerhans cell ADAM17 to promote photosensitivity in lupus
Abstract
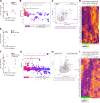
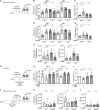
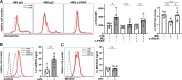
The autoimmune disease lupus erythematosus (lupus) is characterized by photosensitivity, where even ambient ultraviolet radiation (UVR) exposure can lead to development of inflammatory skin lesions. We have previously shown that Langerhans cells (LCs) limit keratinocyte apoptosis and photosensitivity via a disintegrin and metalloprotease 17 (ADAM17)-mediated release of epidermal growth factor receptor (EGFR) ligands and that LC ADAM17 sheddase activity is reduced in lupus. Here, we sought to understand how the lupus skin environment contributes to LC ADAM17 dysfunction and, in the process, differentiate between effects on LC ADAM17 sheddase function, LC ADAM17 expression, and LC numbers. We show through transcriptomic analysis a shared IFN-rich environment in non-lesional skin across human lupus and three murine models: MRL/lpr, B6.Sle1yaa, and imiquimod (IMQ) mice. IFN-I inhibits LC ADAM17 sheddase activity in murine and human LCs, and IFNAR blockade in lupus model mice restores LC ADAM17 sheddase activity, all without consistent effects on LC ADAM17 protein expression or LC numbers. Anti-IFNAR-mediated LC ADAM17 sheddase function restoration is associated with reduced photosensitive responses that are dependent on EGFR signaling and LC ADAM17. Reactive oxygen species (ROS) is a known mediator of ADAM17 activity; we show that UVR-induced LC ROS production is reduced in lupus model mice, restored by anti-IFNAR, and is cytoplasmic in origin. Our findings suggest that IFN-I promotes photosensitivity at least in part by inhibiting UVR-induced LC ADAM17 sheddase function and raise the possibility that anifrolumab ameliorates lupus skin disease in part by restoring this function. This work provides insight into IFN-I-mediated disease mechanisms, LC regulation, and a potential mechanism of action for anifrolumab in lupus.
Keywords: ADAM17; human; immunology; inflammation; interferons; langerhans cells; lupus; mouse; photosensitivity.
© 2024, Li, Zyulina, Seltzer et al.
Conflict of interest statement
TL, VZ, ES, MD, YC, KV, DO, PC, YL, WS, ST, KO, MR, IR No competing interests declared, AD, PL is an employee of AMPEL BioSolutions, but has no financial conflicts of interest to report, NS was awarded the Lupus Therapeutics: The Clinical Trial Network Infrastructure Grant, received by The Albert Einstein College of Medicine. The author received payment for lectures at the Congress of Clinical Rheumatology East and the Congress of Clinical Rheumatology West. The author has no other competing interests to declare, JL has received the grants F31 NIH GM136144 and T32 NIH GM008539. The author has received stock or stock options from NASDAQ/NYSE Ticker: FULC, ABCL, AVXL, VOR, MRNA, BNTX, SAVA, OCGN, CTMX, BCEL, GE. The author has no other competing interests to declare, WA received support for travel and attending Lupus 21st century meeting in 2021. The author has no other competing interests to declare, JZ received a grant from NIH NIAMS, and consulting fees from Hoth Therapeutics and Pfizer. The author received payment for participation on a Data Safety Monitoring Board/ Advisory Board for Hoth Therapeutics and acts as President elect for PASPCR. The author holds stock options from Hoth Therapeutics, FoxWayne Inc and YouV labs. The author has no other competing interests to declare, JK has received grant support from AbbVie, Akros, Allergan, Amgen, Avillion, Biogen, Botanix, Boehringer Ingelheim, Bristol-Myers Squibb, Exicure, Innovaderm, Incyte, Janssen, Kyowa Kirin, Lilly, Nimbus Lackshmi, Novan, Novartis, PAREXEL, Pfizer, Regeneron, UCB, Vitae Pharmaceuticals. The author received consulting fees from AbbVie, Aclaris, Allergan, Almirall, Amgen, Artax Biopharma, Arena, Aristea, Asana, Aurigene, Biogen Idec, Boehringer Ingelheim, Bristol-Myers Squibb, Escalier, Galapagos, Janssen, Kyowa Kirin, Lilly, MoonLake Immunotherapeutics, Nimbus, Novartis, Pfizer, Sanofi, Sienna Biopharmaceuticals, Sun Pharma, Target-Derm, UCB, Valeant, Ventyx. The author has no other competing interests to declare, NA has received the following grants: NIAMS AR080436-01, NIAMS R56AR078686-01 and NIH NIAMS 5R01 GRANT AR070234-05. The author received consulting fees from Immunitas, Shennon Bio and Janssen. The author received payment as a lecturer from 23 and me, Cellino and Bristol Meyer Squibb Genomics. They are also a board member of the Society of Investigative Dermatology. The author has no other competing interests to declare, AJ has received grants from the NIH and the VA and consulting fees from Pfizer. The author has no other competing interests to declare, CB The patent number is US10024844B2 and the title of the patent is "Identification of an inhibitor of iRhom1 or an inhibitor of iRhom2", which is also what the patent relates to. Carl Blobel and the Hospital for Special Surgery have identified iRhom2 inhibitors and have co-founded the start-up company SciRhom in Munich to commercialize these inhibitors, TL has received the following grants: NIH R01AI079178, NIH R21 AR081493, Department of Defense W81XWH-21-LRP-IPA, Lupus Research Alliance Lupus Innovation Award grant, Barbara Volcker Center for Women and Rheumatic Diseases grant. She has also received funding support from the St. Giles Foundation and A Lasting Mark Foundation. She has received consulting fees from Pfizer, and has a received payment from Bristol Meyers Squibb for giving a lecture. The author has received payment for attending Lupus 21st Century meeting. The author has no other competing interests to declare
Figures









Update of
- doi: 10.1101/2021.08.18.456792
References
-
- Ambler WG, Howlander M, Chalasani MLS, Seltzer ES, Sim J, Shin J, Schwartz N, Shipman WD, Dasoveanu D, Carballo CB, Sevim E, Siddique S, Rodeo S, Erkan D, Kataru RP, Mehrara B, Lu TT. Lymphatic Dysfunction in Lupus Contributes to Cutaneous Photosensitivity and Lymph Node B Cell Responses. bioRxiv. 2022 doi: 10.1101/2022.06.13.495930. - DOI
-
- Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. PNAS. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. - DOI - PMC - PubMed
-
- Benci JL, Johnson LR, Choa R, Xu Y, Qiu J, Zhou Z, Xu B, Ye D, Nathanson KL, June CH, Wherry EJ, Zhang NR, Ishwaran H, Hellmann MD, Wolchok JD, Kambayashi T, Minn AJ. Opposing functions of interferon coordinate adaptive and innate immune responses to cancer immune checkpoint blockade. Cell. 2019;178:933–948. doi: 10.1016/j.cell.2019.07.019. - DOI - PMC - PubMed
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
Grants and funding
- R21 AR081493/AR/NIAMS NIH HHS/United States
- R01AR077194/GF/NIH HHS/United States
- R35GM134907/GF/NIH HHS/United States
- R01 AR080436/AR/NIAMS NIH HHS/United States
- R21 AR081493/GF/NIH HHS/United States
- R01 AR077194/AR/NIAMS NIH HHS/United States
- T32 AR071302/AR/NIAMS NIH HHS/United States
- DK099087/GF/NIH HHS/United States
- I01 BX004907/BX/BLRD VA/United States
- K08 AR069111/GF/NIH HHS/United States
- W81XWH-21-LRP-IPA/DOD
- R01 AI079178/AI/NIAID NIH HHS/United States
- T32AR071302/GF/NIH HHS/United States
- R01 DK099087/DK/NIDDK NIH HHS/United States
- K08 AR069111/AR/NIAMS NIH HHS/United States
- R01AI079178/GF/NIH HHS/United States
- J 4638-B FWF/Erwin Schrodinger Fellowship
- T32GM007739/NIH MSTP grant
- R35 GM134907/GM/NIGMS NIH HHS/United States
- S10 OD019986/OD/NIH HHS/United States
- T32 GM007739/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

