Activation of alveolar epithelial ER stress by β-coronavirus infection disrupts surfactant homeostasis in mice: implications for COVID-19 respiratory failure
- PMID: 38860845
- PMCID: PMC11444511
- DOI: 10.1152/ajplung.00324.2023
Activation of alveolar epithelial ER stress by β-coronavirus infection disrupts surfactant homeostasis in mice: implications for COVID-19 respiratory failure
Abstract
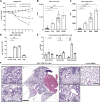
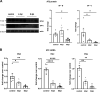
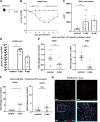
COVID-19 syndrome is characterized by acute lung injury, hypoxemic respiratory failure, and high mortality. Alveolar type 2 (AT2) cells are essential for gas exchange, repair, and regeneration of distal lung epithelium. We have shown that the causative agent, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), and other members of the β-coronavirus genus induce an endoplasmic reticulum (ER) stress response in vitro; however, the consequences for host AT2 cell function in vivo are less understood. To study this, two murine models of coronavirus infection were used-mouse hepatitis virus-1 (MHV-1) in A/J mice and a mouse-adapted SARS-CoV-2 strain. MHV-1-infected mice exhibited dose-dependent weight loss with histological evidence of distal lung injury accompanied by elevated bronchoalveolar lavage fluid (BALF) cell counts and total protein. AT2 cells showed evidence of both viral infection and increased BIP/GRP78 expression, consistent with activation of the unfolded protein response (UPR). The AT2 UPR included increased inositol-requiring enzyme 1α (IRE1α) signaling and a biphasic response in PKR-like ER kinase (PERK) signaling accompanied by marked reductions in AT2 and BALF surfactant protein (SP-B and SP-C) content, increases in surfactant surface tension, and emergence of a reprogrammed epithelial cell population (Krt8+ and Cldn4+). The loss of a homeostatic AT2 cell state was attenuated by treatment with the IRE1α inhibitor OPK-711. As a proof-of-concept, C57BL6 mice infected with mouse-adapted SARS-CoV-2 demonstrated similar lung injury and evidence of disrupted surfactant homeostasis. We conclude that lung injury from β-coronavirus infection results from an aberrant host response, activating multiple AT2 UPR stress pathways, altering surfactant metabolism/function, and changing AT2 cell state, offering a mechanistic link between SARS-CoV-2 infection, AT2 cell biology, and acute respiratory failure.NEW & NOTEWORTHY COVID-19 syndrome is characterized by hypoxemic respiratory failure and high mortality. In this report, we use two murine models to show that β-coronavirus infection produces acute lung injury, which results from an aberrant host response, activating multiple epithelial endoplasmic reticular stress pathways, disrupting pulmonary surfactant metabolism and function, and forcing emergence of an aberrant epithelial transition state. Our results offer a mechanistic link between SARS-CoV-2 infection, AT2 cell biology, and respiratory failure.
Keywords: SARS-CoV-2; alveolar type 2 cell; mouse hepatitis virus-1 (MHV-1); pulmonary surfactant; transitional epithelial cell state.
Conflict of interest statement
No conflicts of interest, financial or otherwise, are declared by the authors.
Figures





Similar articles
-
The IRE1α-XBP1 arm of the unfolded protein response is a host factor activated in SARS-CoV-2 infection.Biochim Biophys Acta Mol Basis Dis. 2024 Jun;1870(5):167193. doi: 10.1016/j.bbadis.2024.167193. Epub 2024 Apr 20. Biochim Biophys Acta Mol Basis Dis. 2024. PMID: 38648902 Free PMC article.
-
Effects of immunoproteasome inhibition on acute respiratory infection with murine hepatitis virus strain 1.J Virol. 2024 Dec 17;98(12):e0123824. doi: 10.1128/jvi.01238-24. Epub 2024 Nov 7. J Virol. 2024. PMID: 39508578 Free PMC article.
-
SARS-CoV-2 ORF3a induces COVID-19-associated kidney injury through HMGB1-mediated cytokine production.mBio. 2024 Nov 13;15(11):e0230824. doi: 10.1128/mbio.02308-24. Epub 2024 Sep 30. mBio. 2024. PMID: 39345136 Free PMC article.
-
Antibody tests for identification of current and past infection with SARS-CoV-2.Cochrane Database Syst Rev. 2022 Nov 17;11(11):CD013652. doi: 10.1002/14651858.CD013652.pub2. Cochrane Database Syst Rev. 2022. PMID: 36394900 Free PMC article.
-
Physical interventions to interrupt or reduce the spread of respiratory viruses.Cochrane Database Syst Rev. 2023 Jan 30;1(1):CD006207. doi: 10.1002/14651858.CD006207.pub6. Cochrane Database Syst Rev. 2023. PMID: 36715243 Free PMC article.
Cited by
-
Idiopathic Pulmonary Fibrosis Caused by Damaged Mitochondria and Imbalanced Protein Homeostasis in Alveolar Epithelial Type II Cell.Adv Biol (Weinh). 2025 Apr;9(4):e2400297. doi: 10.1002/adbi.202400297. Epub 2024 Oct 10. Adv Biol (Weinh). 2025. PMID: 39390651 Free PMC article. Review.
-
Impaired AMPK control of alveolar epithelial cell metabolism promotes pulmonary fibrosis.JCI Insight. 2025 Jul 1;10(15):e182578. doi: 10.1172/jci.insight.182578. eCollection 2025 Aug 8. JCI Insight. 2025. PMID: 40591409 Free PMC article.
References
-
- Mulugeta S, Nureki S, Beers MF. Lost after translation: insights from pulmonary surfactant for understanding the role of alveolar epithelial dysfunction and cellular quality control in fibrotic lung disease. Am J Physiol Lung Cell Mol Physiol 309: L507–L525, 2015. doi: 10.1152/ajplung.00139.2015. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

