Prevalence and diversity of TAL effector-like proteins in fungal endosymbiotic Mycetohabitans spp
- PMID: 38860878
- PMCID: PMC11261895
- DOI: 10.1099/mgen.0.001261
Prevalence and diversity of TAL effector-like proteins in fungal endosymbiotic Mycetohabitans spp
Abstract
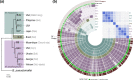
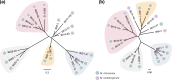
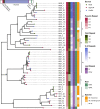
Endofungal Mycetohabitans (formerly Burkholderia) spp. rely on a type III secretion system to deliver mostly unidentified effector proteins when colonizing their host fungus, Rhizopus microsporus. The one known secreted effector family from Mycetohabitans consists of homologues of transcription activator-like (TAL) effectors, which are used by plant pathogenic Xanthomonas and Ralstonia spp. to activate host genes that promote disease. These 'Burkholderia TAL-like (Btl)' proteins bind corresponding specific DNA sequences in a predictable manner, but their genomic target(s) and impact on transcription in the fungus are unknown. Recent phenotyping of Btl mutants of two Mycetohabitans strains revealed that the single Btl in one Mycetohabitans endofungorum strain enhances fungal membrane stress tolerance, while others in a Mycetohabitans rhizoxinica strain promote bacterial colonization of the fungus. The phenotypic diversity underscores the need to assess the sequence diversity and, given that sequence diversity translates to DNA targeting specificity, the functional diversity of Btl proteins. Using a dual approach to maximize capture of Btl protein sequences for our analysis, we sequenced and assembled nine Mycetohabitans spp. genomes using long-read PacBio technology and also mined available short-read Illumina fungal-bacterial metagenomes. We show that btl genes are present across diverse Mycetohabitans strains from Mucoromycota fungal hosts yet vary in sequences and predicted DNA binding specificity. Phylogenetic analysis revealed distinct clades of Btl proteins and suggested that Mycetohabitans might contain more species than previously recognized. Within our data set, Btl proteins were more conserved across M. rhizoxinica strains than across M. endofungorum, but there was also evidence of greater overall strain diversity within the latter clade. Overall, the results suggest that Btl proteins contribute to bacterial-fungal symbioses in myriad ways.
Keywords: Mycetohabitans; Rhizopus; effectors; endofungal bacteria; long-read sequencing; meta-assembled genomes.
Conflict of interest statement
J.E.S. was a paid consultant for Zymergen, Sincarne and Michroma.
Figures



Similar articles
-
A TAL effector-like protein of an endofungal bacterium increases the stress tolerance and alters the transcriptome of the host.Proc Natl Acad Sci U S A. 2020 Jul 21;117(29):17122-17129. doi: 10.1073/pnas.2003857117. Epub 2020 Jul 6. Proc Natl Acad Sci U S A. 2020. PMID: 32632014 Free PMC article.
-
Transcription activator-like effectors from endosymbiotic bacteria control the reproduction of their fungal host.mBio. 2023 Dec 19;14(6):e0182423. doi: 10.1128/mbio.01824-23. Epub 2023 Nov 16. mBio. 2023. PMID: 37971247 Free PMC article.
-
Deazaflavin metabolite produced by endosymbiotic bacteria controls fungal host reproduction.ISME J. 2024 Jan 8;18(1):wrae074. doi: 10.1093/ismejo/wrae074. ISME J. 2024. PMID: 38691425 Free PMC article.
-
TAL effector-DNA specificity.Virulence. 2010 Sep-Oct;1(5):428-32. doi: 10.4161/viru.1.5.12863. Virulence. 2010. PMID: 21178484 Review.
-
TAL effectors--pathogen strategies and plant resistance engineering.New Phytol. 2014 Dec;204(4):823-32. doi: 10.1111/nph.13015. New Phytol. 2014. PMID: 25539004 Review.
Cited by
-
Endofungal bacteria as hidden facilitators of biotic interactions.ISME J. 2025 Jan 2;19(1):wraf128. doi: 10.1093/ismejo/wraf128. ISME J. 2025. PMID: 40581745 Free PMC article. Review.
References
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

