Single-tissue proteomics in Caenorhabditis elegans reveals proteins resident in intestinal lysosome-related organelles
- PMID: 38861598
- PMCID: PMC11194598
- DOI: 10.1073/pnas.2322588121
Single-tissue proteomics in Caenorhabditis elegans reveals proteins resident in intestinal lysosome-related organelles
Abstract
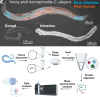
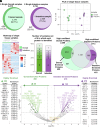
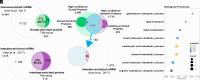
The nematode intestine is the primary site for nutrient uptake and storage as well as the synthesis of biomolecules; lysosome-related organelles known as gut granules are important for many of these functions. Aspects of intestine biology are not well understood, including the export of the nutrients it imports and the molecules it synthesizes, as well as the complete functions and protein content of the gut granules. Here, we report a mass spectrometry (MS)-based proteomic analysis of the intestine of the Caenorhabditis elegans and of its gut granules. Overall, we identified approximately 5,000 proteins each in the intestine and the gonad and showed that most of these proteins can be detected in samples extracted from a single worm, suggesting the feasibility of individual-level genetic analysis using proteomes. Comparing proteomes and published transcriptomes of the intestine and the gonad, we identified proteins that appear to be synthesized in the intestine and then transferred to the gonad. To identify gut granule proteins, we compared the proteome of individual intestines deficient in gut granules to the wild type. The identified gut granule proteome includes proteins known to be exclusively localized to the granules and additional putative gut granule proteins. We selected two of these putative gut granule proteins for validation via immunohistochemistry, and our successful confirmation of both suggests that our strategy was effective in identifying the gut granule proteome. Our results demonstrate the practicability of single-tissue MS-based proteomic analysis in small organisms and in its future utility.
Keywords: lysosome-related organelle; microproteomics; tissue-specific mass-spectrometry; yolk protein.
Conflict of interest statement
Competing interests statement:The authors declare no competing interest.
Figures



Similar articles
-
Roles of CUP-5, the Caenorhabditis elegans orthologue of human TRPML1, in lysosome and gut granule biogenesis.BMC Cell Biol. 2010 Jun 11;11:40. doi: 10.1186/1471-2121-11-40. BMC Cell Biol. 2010. PMID: 20540742 Free PMC article.
-
Asymmetric organelle positioning during epithelial polarization of C. elegans intestinal cells.Dev Biol. 2022 Jan;481:75-94. doi: 10.1016/j.ydbio.2021.09.007. Epub 2021 Sep 29. Dev Biol. 2022. PMID: 34597675 Free PMC article.
-
Function of the Caenorhabditis elegans ABC transporter PGP-2 in the biogenesis of a lysosome-related fat storage organelle.Mol Biol Cell. 2007 Mar;18(3):995-1008. doi: 10.1091/mbc.e06-08-0685. Epub 2007 Jan 3. Mol Biol Cell. 2007. PMID: 17202409 Free PMC article.
-
Proteomics in Caenorhabditis elegans.Brief Funct Genomic Proteomic. 2008 May;7(3):205-10. doi: 10.1093/bfgp/eln014. Epub 2008 Mar 27. Brief Funct Genomic Proteomic. 2008. PMID: 18372286 Review.
-
Mass spectrometry-based shotgun proteomic analysis of C. elegans protein complexes.WormBook. 2014 Jun 24:1-18. doi: 10.1895/wormbook.1.171.1. WormBook. 2014. PMID: 24967700 Free PMC article. Review.
Cited by
-
The neurotrophic factor MANF regulates autophagy and lysosome function to promote proteostasis in Caenorhabditis elegans.Proc Natl Acad Sci U S A. 2024 Oct 22;121(43):e2403906121. doi: 10.1073/pnas.2403906121. Epub 2024 Oct 17. Proc Natl Acad Sci U S A. 2024. PMID: 39418305 Free PMC article.
-
In-cell processing enables rapid and in-depth proteome analysis of low-input Caenorhabditis elegans.bioRxiv [Preprint]. 2024 Sep 19:2024.09.18.613705. doi: 10.1101/2024.09.18.613705. bioRxiv. 2024. Update in: Anal Chem. 2025 May 6;97(17):9159-9167. doi: 10.1021/acs.analchem.4c05003. PMID: 39345438 Free PMC article. Updated. Preprint.
-
Simple In-Cell Processing Enables Deep Proteome Analysis of Low-Input Caenorhabditis elegans.Anal Chem. 2025 May 6;97(17):9159-9167. doi: 10.1021/acs.analchem.4c05003. Epub 2025 Apr 21. Anal Chem. 2025. PMID: 40258293 Free PMC article.
-
Lipid Oxidation at the Crossroads: Oxidative Stress and Neurodegeneration Explored in Caenorhabditis elegans.Antioxidants (Basel). 2025 Jan 10;14(1):78. doi: 10.3390/antiox14010078. Antioxidants (Basel). 2025. PMID: 39857412 Free PMC article. Review.
-
Detecting gene expression in Caenorhabditis elegans.Genetics. 2025 Jan 8;229(1):1-108. doi: 10.1093/genetics/iyae167. Genetics. 2025. PMID: 39693264 Free PMC article. Review.
References
-
- Sulston J. E., Horvitz H. R., Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 56, 110–156 (1977). - PubMed
-
- Hedgecock E. M., White J. G., Polyploid tissues in the nematode Caenorhabditis elegans. Dev. Biol. 107, 128–133 (1985). - PubMed
-
- Ashrafi K., et al. , Genome-wide RNAi analysis of Caenorhabditis elegans fat regulatory genes. Nature 421, 268–272 (2003). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

