The non-catalytic domains of O-GlcNAc cycling enzymes present new opportunities for function-specific control
- PMID: 38861851
- PMCID: PMC11323188
- DOI: 10.1016/j.cbpa.2024.102476
The non-catalytic domains of O-GlcNAc cycling enzymes present new opportunities for function-specific control
Abstract
O-GlcNAcylation is an essential protein glycosylation governed by two O-GlcNAc cycling enzymes: O-GlcNAc transferase (OGT) installs a single sugar moiety N-acetylglucosamine (GlcNAc) on protein serine and threonine residues, and O-GlcNAcase (OGA) removes them. Aberrant O-GlcNAcylation has been implicated in various diseases. However, the large repertoire of more than 1000 O-GlcNAcylated proteins and the elusive mechanisms of OGT/OGA in substrate recognition present significant challenges in targeting the dysregulated O-GlcNAcylation for therapeutic development. Recently, emerging evidence suggested that the non-catalytic domains play critical roles in regulating the functional specificity of OGT/OGA via modulating their protein interactions and substrate recognition. Here, we discuss recent studies on the structures, mechanisms, and related tools of the OGT/OGA non-catalytic domains, highlighting new opportunities for function-specific control.
Keywords: Function-specific modulator; Non-catalytic domain; O-GlcNAc transferase (OGT); O-GlcNAcase (OGA).
Copyright © 2024 Elsevier Ltd. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures
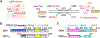

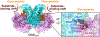
Similar articles
-
Elucidating the protein substrate recognition of O-GlcNAc transferase (OGT) toward O-GlcNAcase (OGA) using a GlcNAc electrophilic probe.Int J Biol Macromol. 2021 Feb 1;169:51-59. doi: 10.1016/j.ijbiomac.2020.12.078. Epub 2020 Dec 18. Int J Biol Macromol. 2021. PMID: 33333092 Free PMC article.
-
Enhancing protein O-GlcNAcylation in down syndrome mice mitigates memory dysfunctions through the rescue of mitochondrial bioenergetics, stress responses and pathological markers.Redox Biol. 2025 Sep;85:103769. doi: 10.1016/j.redox.2025.103769. Epub 2025 Jul 16. Redox Biol. 2025. PMID: 40684658 Free PMC article.
-
Protein O-GlcNAcylation in reproductive biology and the impact of metabolic disease.Hum Reprod Update. 2025 Jun 26:dmaf013. doi: 10.1093/humupd/dmaf013. Online ahead of print. Hum Reprod Update. 2025. PMID: 40574323
-
Chemical and Biochemical Strategies To Explore the Substrate Recognition of O-GlcNAc-Cycling Enzymes.Chembiochem. 2019 Feb 1;20(3):312-318. doi: 10.1002/cbic.201800481. Epub 2018 Nov 12. Chembiochem. 2019. PMID: 30199580 Free PMC article. Review.
-
Molecular Interrogation to Crack the Case of O-GlcNAc.Chemistry. 2020 Sep 21;26(53):12086-12100. doi: 10.1002/chem.202000155. Epub 2020 Jul 20. Chemistry. 2020. PMID: 32207184 Free PMC article. Review.
Cited by
-
Evidence for Functional Regulation of the KLHL3/WNK Pathway by O-GlcNAcylation.bioRxiv [Preprint]. 2025 Feb 27:2025.02.27.640596. doi: 10.1101/2025.02.27.640596. bioRxiv. 2025. Update in: Glycobiology. 2025 Aug 11;35(9):cwaf046. doi: 10.1093/glycob/cwaf046. PMID: 40060460 Free PMC article. Updated. Preprint.
References
-
- Zachara NE, Akimoto Y, Boyce M, Hart GW: The O-GlcNAc modification. In Essentials of Glycobiology [Internet]. Ch. 19. Edited by Varki A, Cummings RD, Esko JD, Stanley P, Hart GW, Aebi M, Mohnen D, Kinoshita T, Packer NH, Prestegard JH, et al. Cold Spring Harbor Laboratory Press; 2022. - PubMed
-
- Zou Y, Pei J, Long H, Lan L, Dong K, Wang T, Li M, Zhao Z, Zhu L, Zhang G, et al. : H4S47 O-GlcNAcylation regulates the activation of mammalian replication origins. Nat Struct Mol Biol 2023, 30:800–811. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

