Structural and functional insights into the C-terminal signal domain of the Bacteroidetes type-IX secretion system
- PMID: 38862016
- PMCID: PMC11285876
- DOI: 10.1098/rsob.230448
Structural and functional insights into the C-terminal signal domain of the Bacteroidetes type-IX secretion system
Abstract
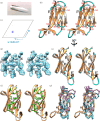
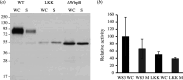
Gram-negative bacteria from the Bacteroidota phylum possess a type-IX secretion system (T9SS) for protein secretion, which requires cargoes to have a C-terminal domain (CTD). Structurally analysed CTDs are from Porphyromonas gingivalis proteins RgpB, HBP35, PorU and PorZ, which share a compact immunoglobulin-like antiparallel 3+4 β-sandwich (β1-β7). This architecture is essential as a P. gingivalis strain with a single-point mutant of RgpB disrupting the interaction of the CTD with its preceding domain prevented secretion of the protein. Next, we identified the C-terminus ('motif C-t.') and the loop connecting strands β3 and β4 ('motif Lβ3β4') as conserved. We generated two strains with insertion and replacement mutants of PorU, as well as three strains with ablation and point mutants of RgpB, which revealed both motifs to be relevant for T9SS function. Furthermore, we determined the crystal structure of the CTD of mirolase, a cargo of the Tannerella forsythia T9SS, which shares the same general topology as in Porphyromonas CTDs. However, motif Lβ3β4 was not conserved. Consistently, P. gingivalis could not properly secrete a chimaeric protein with the CTD of peptidylarginine deiminase replaced with this foreign CTD. Thus, the incompatibility of the CTDs between these species prevents potential interference between their T9SSs.
Keywords: T9SS; X-ray crystal structure; bacterial virulence factor; infectious disease; periodontal disease; protein secretion.
Conflict of interest statement
We declare we have no competing interests.
Figures
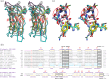

Similar articles
-
The outer-membrane export signal of Porphyromonas gingivalis type IX secretion system (T9SS) is a conserved C-terminal β-sandwich domain.Sci Rep. 2016 Mar 23;6:23123. doi: 10.1038/srep23123. Sci Rep. 2016. PMID: 27005013 Free PMC article.
-
PorZ, an Essential Component of the Type IX Secretion System of Porphyromonas gingivalis, Delivers Anionic Lipopolysaccharide to the PorU Sortase for Transpeptidase Processing of T9SS Cargo Proteins.mBio. 2021 Feb 23;12(1):e02262-20. doi: 10.1128/mBio.02262-20. mBio. 2021. PMID: 33622730 Free PMC article.
-
Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis.Sci Rep. 2016 Nov 24;6:37708. doi: 10.1038/srep37708. Sci Rep. 2016. PMID: 27883039 Free PMC article.
-
The Type IX Secretion System (T9SS): Highlights and Recent Insights into Its Structure and Function.Front Cell Infect Microbiol. 2017 May 26;7:215. doi: 10.3389/fcimb.2017.00215. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28603700 Free PMC article. Review.
-
Type IX secretion: the generation of bacterial cell surface coatings involved in virulence, gliding motility and the degradation of complex biopolymers.Mol Microbiol. 2017 Oct;106(1):35-53. doi: 10.1111/mmi.13752. Epub 2017 Aug 9. Mol Microbiol. 2017. PMID: 28714554 Review.
Cited by
-
C-terminal glycosylation of type IX secretion system cargo proteins in Prevotella intermedia with both short and long secretion signals.Open Biol. 2025 Mar;15(3):240335. doi: 10.1098/rsob.240335. Epub 2025 Mar 26. Open Biol. 2025. PMID: 40132644 Free PMC article.
-
Exploring heme and iron acquisition strategies of Porphyromonas gingivalis-current facts and hypotheses.FEMS Microbiol Rev. 2025 Jan 14;49:fuaf019. doi: 10.1093/femsre/fuaf019. FEMS Microbiol Rev. 2025. PMID: 40343779 Free PMC article. Review.
-
Defining the role of Hmu and Hus systems in Porphyromonas gingivalis heme and iron homeostasis and virulence.Sci Rep. 2024 Dec 28;14(1):31156. doi: 10.1038/s41598-024-82326-6. Sci Rep. 2024. PMID: 39730829 Free PMC article.
-
PorA of the Type IX Secretion Is a Ligand of the PorXY Two-Component Regulatory System in Porphyromonas gingivalis.Mol Microbiol. 2025 Jun;123(6):569-585. doi: 10.1111/mmi.15363. Epub 2025 Apr 8. Mol Microbiol. 2025. PMID: 40195800 Free PMC article.
-
Dissecting Cytophagalysin: Structural and Biochemical Studies of a Bacterial Pappalysin-Family Metallopeptidase.Biomolecules. 2024 Dec 16;14(12):1604. doi: 10.3390/biom14121604. Biomolecules. 2024. PMID: 39766312 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

