Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins
- PMID: 38862021
- PMCID: PMC11286163
- DOI: 10.1098/rsob.240025
Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins
Abstract
Faithful transmission of genetic material is crucial for the survival of all organisms. In many eukaryotes, a feedback control mechanism called the spindle checkpoint ensures chromosome segregation fidelity by delaying cell cycle progression until all chromosomes achieve proper attachment to the mitotic spindle. Kinetochores are the macromolecular complexes that act as the interface between chromosomes and spindle microtubules. While most eukaryotes have canonical kinetochore proteins that are widely conserved, kinetoplastids such as Trypanosoma brucei have a seemingly unique set of kinetochore proteins including KKT1-25. It remains poorly understood how kinetoplastids regulate cell cycle progression or ensure chromosome segregation fidelity. Here, we report a crystal structure of the C-terminal domain of KKT14 from Apiculatamorpha spiralis and uncover that it is a pseudokinase. Its structure is most similar to the kinase domain of a spindle checkpoint protein Bub1. In addition, KKT14 has a putative ABBA motif that is present in Bub1 and its paralogue BubR1. We also find that the N-terminal part of KKT14 interacts with KKT15, whose WD40 repeat beta-propeller is phylogenetically closely related to a direct interactor of Bub1/BubR1 called Bub3. Our findings indicate that KKT14-KKT15 are divergent orthologues of Bub1/BubR1-Bub3, which promote accurate chromosome segregation in trypanosomes.
Keywords: Trypanosoma brucei; chromosome segregation; kinetochore; kinetoplastid; spindle checkpoint.
Conflict of interest statement
We declare we have no competing interests.
Figures

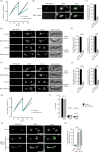
![KKT14 has a putative ABBA motif. Schematic of the T. brucei KKT14 protein and a multiple sequence alignment showing a putative ABBA motif (consensus Fx[ILV][FHY]x[DE]](https://cdn.ncbi.nlm.nih.gov/pmc/blobs/e4be/11286163/c7c8c4fe4846/rsob.240025.f002.gif)



Similar articles
-
Aurora B controls anaphase onset and error-free chromosome segregation in trypanosomes.J Cell Biol. 2024 Nov 4;223(11):e202401169. doi: 10.1083/jcb.202401169. Epub 2024 Aug 28. J Cell Biol. 2024. PMID: 39196069 Free PMC article.
-
BuGZ facilitates loading of spindle assembly checkpoint proteins to kinetochores in early mitosis.J Biol Chem. 2020 Oct 23;295(43):14666-14677. doi: 10.1074/jbc.RA120.013598. Epub 2020 Aug 20. J Biol Chem. 2020. PMID: 32820050 Free PMC article.
-
Role of Intrinsic and Extrinsic Factors in the Regulation of the Mitotic Checkpoint Kinase Bub1.PLoS One. 2015 Dec 10;10(12):e0144673. doi: 10.1371/journal.pone.0144673. eCollection 2015. PLoS One. 2015. PMID: 26658523 Free PMC article.
-
Bub1 and BubR1: at the interface between chromosome attachment and the spindle checkpoint.Mol Cell Biol. 2011 Aug;31(15):3085-93. doi: 10.1128/MCB.05326-11. Epub 2011 May 31. Mol Cell Biol. 2011. PMID: 21628528 Free PMC article. Review.
-
Evolutionary cell biology of chromosome segregation: insights from trypanosomes.Open Biol. 2013 May 1;3(5):130023. doi: 10.1098/rsob.130023. Open Biol. 2013. PMID: 23635522 Free PMC article. Review.
Cited by
-
An unconventional regulatory circuitry involving Aurora B controls anaphase onset and error-free chromosome segregation in trypanosomes.bioRxiv [Preprint]. 2024 Jan 20:2024.01.20.576407. doi: 10.1101/2024.01.20.576407. bioRxiv. 2024. Update in: J Cell Biol. 2024 Nov 4;223(11):e202401169. doi: 10.1083/jcb.202401169. PMID: 38293145 Free PMC article. Updated. Preprint.
-
On the possibility of yet a third kinetochore system in the protist phylum Euglenozoa.mBio. 2024 Dec 11;15(12):e0293624. doi: 10.1128/mbio.02936-24. Epub 2024 Oct 30. mBio. 2024. PMID: 39475241 Free PMC article.
-
Aurora B controls anaphase onset and error-free chromosome segregation in trypanosomes.J Cell Biol. 2024 Nov 4;223(11):e202401169. doi: 10.1083/jcb.202401169. Epub 2024 Aug 28. J Cell Biol. 2024. PMID: 39196069 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

