A virtual rodent predicts the structure of neural activity across behaviours
- PMID: 38862024
- PMCID: PMC12080270
- DOI: 10.1038/s41586-024-07633-4
A virtual rodent predicts the structure of neural activity across behaviours
Erratum in
-
Author Correction: A virtual rodent predicts the structure of neural activity across behaviours.Nature. 2025 Sep;645(8079):E1. doi: 10.1038/s41586-025-09407-y. Nature. 2025. PMID: 40790097 Free PMC article. No abstract available.
Abstract
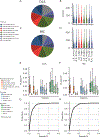
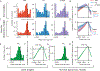
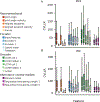
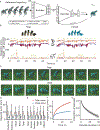
Animals have exquisite control of their bodies, allowing them to perform a diverse range of behaviours. How such control is implemented by the brain, however, remains unclear. Advancing our understanding requires models that can relate principles of control to the structure of neural activity in behaving animals. Here, to facilitate this, we built a 'virtual rodent', in which an artificial neural network actuates a biomechanically realistic model of the rat1 in a physics simulator2. We used deep reinforcement learning3-5 to train the virtual agent to imitate the behaviour of freely moving rats, thus allowing us to compare neural activity recorded in real rats to the network activity of a virtual rodent mimicking their behaviour. We found that neural activity in the sensorimotor striatum and motor cortex was better predicted by the virtual rodent's network activity than by any features of the real rat's movements, consistent with both regions implementing inverse dynamics6. Furthermore, the network's latent variability predicted the structure of neural variability across behaviours and afforded robustness in a way consistent with the minimal intervention principle of optimal feedback control7. These results demonstrate how physical simulation of biomechanically realistic virtual animals can help interpret the structure of neural activity across behaviour and relate it to theoretical principles of motor control.
© 2024. The Author(s), under exclusive licence to Springer Nature Limited.
Figures











References
-
- Merel J et al. Deep neuroethology of a virtual rodent. in Eighth International Conference on Learning Representations (2020).
-
- Todorov E, Erez T & Tassa Y MuJoCo: A physics engine for model-based control. in 2012 IEEE/RSJ International Conference on Intelligent Robots and Systems 5026–5033 (ieeexplore.ieee.org, 2012).
-
- Hasenclever L, Pardo F, Hadsell R, Heess N & Merel J CoMic: Complementary Task Learning & Mimicry for Reusable Skills. 119, 4105–4115 (13–18 Jul 2020).
-
- Merel J et al. Neural Probabilistic Motor Primitives for Humanoid Control. in 7th International Conference on Learning Representations, ICLR 2019, New Orleans, LA, USA, May 6–9, 2019 (OpenReview.net, 2019).
-
- Peng XB, Abbeel P, Levine S & van de Panne M DeepMimic: example-guided deep reinforcement learning of physics-based character skills. ACM Trans. Graph. 37, 1–14 (2018).
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

