Molecular Evolution of Protein Sequences and Codon Usage in Monkeypox Viruses
- PMID: 38862422
- PMCID: PMC11425058
- DOI: 10.1093/gpbjnl/qzad003
Molecular Evolution of Protein Sequences and Codon Usage in Monkeypox Viruses
Abstract
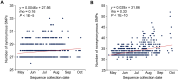
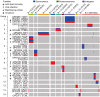
The monkeypox virus (mpox virus, MPXV) epidemic in 2022 has posed a significant public health risk. Yet, the evolutionary principles of MPXV remain largely unknown. Here, we examined the evolutionary patterns of protein sequences and codon usage in MPXV. We first demonstrated the signal of positive selection in OPG027, specifically in the Clade I lineage of MPXV. Subsequently, we discovered accelerated protein sequence evolution over time in the variants responsible for the 2022 outbreak. Furthermore, we showed strong epistasis between amino acid substitutions located in different genes. The codon adaptation index (CAI) analysis revealed that MPXV genes tended to use more non-preferred codons compared to human genes, and the CAI decreased over time and diverged between clades, with Clade I > IIa and IIb-A > IIb-B. While the decrease in fatality rate among the three groups aligned with the CAI pattern, it remains unclear whether this correlation was coincidental or if the deoptimization of codon usage in MPXV led to a reduction in fatality rates. This study sheds new light on the mechanisms that govern the evolution of MPXV in human populations.
Keywords: OPG027; Accelerated evolution; Codon usage bias; Mpox virus; Positive selection.
© The Author(s) 2024. Published by Oxford University Press and Science Press on behalf of the Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Conflict of interest statement
Ke-Jia Shan and Yaling Hu are current employees of Sinovac Biotech Ltd. The other authors have declared no competing interests.
Figures


Similar articles
-
Insights into the Evolution and Host Adaptation of the Monkeypox Virus from a Codon Usage Perspective: Focus on the Ongoing 2022 Outbreak.Int J Mol Sci. 2023 Jul 16;24(14):11524. doi: 10.3390/ijms241411524. Int J Mol Sci. 2023. PMID: 37511283 Free PMC article.
-
Non-adaptive evolution in codon usage of human-origin monkeypox virus.Comp Immunol Microbiol Infect Dis. 2023 Sep;100:102024. doi: 10.1016/j.cimid.2023.102024. Epub 2023 Jul 17. Comp Immunol Microbiol Infect Dis. 2023. PMID: 37487313
-
Evolution of monkeypox virus from 2017 to 2022: In the light of point mutations.Front Microbiol. 2022 Dec 14;13:1037598. doi: 10.3389/fmicb.2022.1037598. eCollection 2022. Front Microbiol. 2022. PMID: 36590408 Free PMC article.
-
Differences in pathogenicity among the mpox virus clades: impact on drug discovery and vaccine development.Trends Pharmacol Sci. 2023 Oct;44(10):719-739. doi: 10.1016/j.tips.2023.08.003. Epub 2023 Sep 4. Trends Pharmacol Sci. 2023. PMID: 37673695 Review.
-
Concurrent Clade I and Clade II Monkeypox Virus Circulation, Cameroon, 1979-2022.Emerg Infect Dis. 2024 Mar;30(3):432-443. doi: 10.3201/eid3003.230861. Epub 2024 Feb 7. Emerg Infect Dis. 2024. PMID: 38325363 Free PMC article. Review.
Cited by
-
Selective events at individual sites underlie the evolution of monkeypox virus clades.Virus Evol. 2023 May 20;9(1):vead031. doi: 10.1093/ve/vead031. eCollection 2023. Virus Evol. 2023. PMID: 37305708 Free PMC article.
-
Evolutionary trajectory and characteristics of Mpox virus in 2023 based on a large-scale genomic surveillance in Shenzhen, China.Nat Commun. 2024 Aug 28;15(1):7452. doi: 10.1038/s41467-024-51737-4. Nat Commun. 2024. PMID: 39198414 Free PMC article.
-
Narciclasine inhibits vaccinia virus infection by activating the RhoA signaling pathway.Biosaf Health. 2024 Nov 19;6(6):341-349. doi: 10.1016/j.bsheal.2024.11.002. eCollection 2024 Dec. Biosaf Health. 2024. PMID: 40078979 Free PMC article.
-
The pathogenicity and multi-organ proteomic profiles of Mpox virus infection in SIVmac239-infected rhesus macaques.Nat Commun. 2025 Aug 17;16(1):7653. doi: 10.1038/s41467-025-62919-z. Nat Commun. 2025. PMID: 40819122 Free PMC article.
-
Phylogenetic Analysis of the Mpox Virus in Sub-Saharan Africa (2022-2024).Biology (Basel). 2025 Jun 26;14(7):773. doi: 10.3390/biology14070773. Biology (Basel). 2025. PMID: 40723332 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

