Substrate and Functional Diversity of Protein Lysine Post-translational Modifications
- PMID: 38862432
- PMCID: PMC12016574
- DOI: 10.1093/gpbjnl/qzae019
Substrate and Functional Diversity of Protein Lysine Post-translational Modifications
Abstract
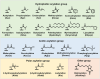
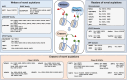
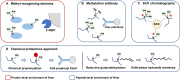
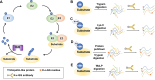
Lysine post-translational modifications (PTMs) are widespread and versatile protein PTMs that are involved in diverse biological processes by regulating the fundamental functions of histone and non-histone proteins. Dysregulation of lysine PTMs is implicated in many diseases, and targeting lysine PTM regulatory factors, including writers, erasers, and readers, has become an effective strategy for disease therapy. The continuing development of mass spectrometry (MS) technologies coupled with antibody-based affinity enrichment technologies greatly promotes the discovery and decoding of PTMs. The global characterization of lysine PTMs is crucial for deciphering the regulatory networks, molecular functions, and mechanisms of action of lysine PTMs. In this review, we focus on lysine PTMs, and provide a summary of the regulatory enzymes of diverse lysine PTMs and the proteomics advances in lysine PTMs by MS technologies. We also discuss the types and biological functions of lysine PTM crosstalks on histone and non-histone proteins and current druggable targets of lysine PTM regulatory factors for disease therapy.
Keywords: Acylation; Drug target; PTM crosstalk; Protein lysine PTM; Regulatory enzyme.
© The Author(s) 2024. Published by Oxford University Press and Science Press on behalf of the Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Conflict of interest statement
The authors have declared no competing interests.
Figures
References
-
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol 2014;15:536–50. - PubMed
-
- Shvedunova M, Akhtar A. Modulation of cellular processes by histone and non-histone protein acetylation. Nat Rev Mol Cell Biol 2022;23:329–49. - PubMed
-
- Janssen SM, Lorincz MC. Interplay between chromatin marks in development and disease. Nat Rev Genet 2022;23:137–53. - PubMed
-
- Hamamoto R, Saloura V, Nakamura Y. Critical roles of non-histone protein lysine methylation in human tumorigenesis. Nat Rev Cancer 2015;15:110–24. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

