Mapping the landscape of histomorphological cancer phenotypes using self-supervised learning on unannotated pathology slides
- PMID: 38862472
- PMCID: PMC11525555
- DOI: 10.1038/s41467-024-48666-7
Mapping the landscape of histomorphological cancer phenotypes using self-supervised learning on unannotated pathology slides
Abstract
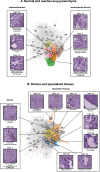
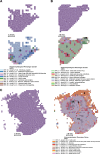
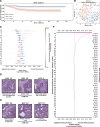
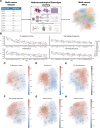
Cancer diagnosis and management depend upon the extraction of complex information from microscopy images by pathologists, which requires time-consuming expert interpretation prone to human bias. Supervised deep learning approaches have proven powerful, but are inherently limited by the cost and quality of annotations used for training. Therefore, we present Histomorphological Phenotype Learning, a self-supervised methodology requiring no labels and operating via the automatic discovery of discriminatory features in image tiles. Tiles are grouped into morphologically similar clusters which constitute an atlas of histomorphological phenotypes (HP-Atlas), revealing trajectories from benign to malignant tissue via inflammatory and reactive phenotypes. These clusters have distinct features which can be identified using orthogonal methods, linking histologic, molecular and clinical phenotypes. Applied to lung cancer, we show that they align closely with patient survival, with histopathologically recognised tumor types and growth patterns, and with transcriptomic measures of immunophenotype. These properties are maintained in a multi-cancer study.
© 2024. The Author(s).
Conflict of interest statement
The authors declare the following competing interests: A.T. is a co-founder of Imagenomix; N.C. is a scientific advisor for Imagenomix. The other authors declare that they have no competing interests.
Figures





Similar articles
-
Weakly Supervised Deep Learning for Whole Slide Lung Cancer Image Analysis.IEEE Trans Cybern. 2020 Sep;50(9):3950-3962. doi: 10.1109/TCYB.2019.2935141. Epub 2019 Sep 2. IEEE Trans Cybern. 2020. PMID: 31484154
-
Self-supervised learning reveals clinically relevant histomorphological patterns for therapeutic strategies in colon cancer.Nat Commun. 2025 Mar 8;16(1):2328. doi: 10.1038/s41467-025-57541-y. Nat Commun. 2025. PMID: 40057490 Free PMC article.
-
Self-Supervised Learning Reveals Clinically Relevant Histomorphological Patterns for Therapeutic Strategies in Colon Cancer.bioRxiv [Preprint]. 2024 Mar 21:2024.02.26.582106. doi: 10.1101/2024.02.26.582106. bioRxiv. 2024. Update in: Nat Commun. 2025 Mar 08;16(1):2328. doi: 10.1038/s41467-025-57541-y. PMID: 38496571 Free PMC article. Updated. Preprint.
-
Towards label-efficient automatic diagnosis and analysis: a comprehensive survey of advanced deep learning-based weakly-supervised, semi-supervised and self-supervised techniques in histopathological image analysis.Phys Med Biol. 2022 Oct 14;67(20). doi: 10.1088/1361-6560/ac910a. Phys Med Biol. 2022. PMID: 36084627 Review.
-
Unsupervised and self-supervised deep learning approaches for biomedical text mining.Brief Bioinform. 2021 Mar 22;22(2):1592-1603. doi: 10.1093/bib/bbab016. Brief Bioinform. 2021. PMID: 33569575 Review.
Cited by
-
Deep learning model for automated diagnosis of moyamoya disease based on magnetic resonance angiography.EClinicalMedicine. 2024 Nov 5;77:102888. doi: 10.1016/j.eclinm.2024.102888. eCollection 2024 Nov. EClinicalMedicine. 2024. PMID: 39559186 Free PMC article.
-
Integrated multicenter deep learning system for prognostic prediction in bladder cancer.NPJ Precis Oncol. 2024 Oct 16;8(1):233. doi: 10.1038/s41698-024-00731-6. NPJ Precis Oncol. 2024. PMID: 39414931 Free PMC article.
-
The Histomorphology to Molecular Transition: Exploring the Genomic Landscape of Poorly Differentiated Epithelial Endometrial Cancers.Cells. 2025 Mar 5;14(5):382. doi: 10.3390/cells14050382. Cells. 2025. PMID: 40072110 Free PMC article. Review.
-
Artificial intelligence in lung cancer: current applications, future perspectives, and challenges.Front Oncol. 2024 Dec 23;14:1486310. doi: 10.3389/fonc.2024.1486310. eCollection 2024. Front Oncol. 2024. PMID: 39763611 Free PMC article. Review.
-
Systematic scoping review of external validation studies of AI pathology models for lung cancer diagnosis.NPJ Precis Oncol. 2025 Jun 7;9(1):166. doi: 10.1038/s41698-025-00940-7. NPJ Precis Oncol. 2025. PMID: 40483288 Free PMC article.
References
-
- Almendro, V., Marusyk, A. & Polyak, K. Cellular heterogeneity and molecular evolution in cancer. Annu. Rev. Pathol. Mech. Dis.8, 277–302 (2013). - PubMed
-
- de Sousa, V. M. L. & Carvalho, L. Heterogeneity in lung cancer. Pathobiology85, 96–107 (2018). - PubMed
-
- Kujan, O. et al. Why oral histopathology suffers inter-observer variability on grading oral epithelial dysplasia: an attempt to understand the sources of variation. Oral. Oncol.43, 224–231 (2007). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

