Direct RNA sequencing of astronaut blood reveals spaceflight-associated m6A increases and hematopoietic transcriptional responses
- PMID: 38862496
- PMCID: PMC11166648
- DOI: 10.1038/s41467-024-48929-3
Direct RNA sequencing of astronaut blood reveals spaceflight-associated m6A increases and hematopoietic transcriptional responses
Abstract
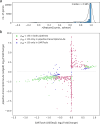
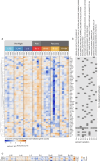
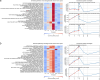
The advent of civilian spaceflight challenges scientists to precisely describe the effects of spaceflight on human physiology, particularly at the molecular and cellular level. Newer, nanopore-based sequencing technologies can quantitatively map changes in chemical structure and expression at single molecule resolution across entire isoforms. We perform long-read, direct RNA nanopore sequencing, as well as Ultima high-coverage RNA-sequencing, of whole blood sampled longitudinally from four SpaceX Inspiration4 astronauts at seven timepoints, spanning pre-flight, day of return, and post-flight recovery. We report key genetic pathways, including changes in erythrocyte regulation, stress induction, and immune changes affected by spaceflight. We also present the first m6A methylation profiles for a human space mission, suggesting a significant spike in m6A levels immediately post-flight. These data and results represent the first longitudinal long-read RNA profiles and RNA modification maps for each gene for astronauts, improving our understanding of the human transcriptome's dynamic response to spaceflight.
© 2024. The Author(s).
Conflict of interest statement
C.E.M. is a co-founder of Cosmica Biosciences. The remaining authors declare no competing interests.
Figures


Similar articles
-
The transcriptome response of astronaut leukocytes to long missions aboard the International Space Station reveals immune modulation.Front Immunol. 2023 Jun 22;14:1171103. doi: 10.3389/fimmu.2023.1171103. eCollection 2023. Front Immunol. 2023. PMID: 37426644 Free PMC article.
-
Spaceflight-Associated Brain White Matter Microstructural Changes and Intracranial Fluid Redistribution.JAMA Neurol. 2019 Apr 1;76(4):412-419. doi: 10.1001/jamaneurol.2018.4882. JAMA Neurol. 2019. PMID: 30673793 Free PMC article.
-
Association of Long-Duration Spaceflight With Anterior and Posterior Ocular Structure Changes in Astronauts and Their Recovery.JAMA Ophthalmol. 2020 May 1;138(5):553-559. doi: 10.1001/jamaophthalmol.2020.0673. JAMA Ophthalmol. 2020. PMID: 32239198 Free PMC article.
-
Neurosurgery and spinal adaptations in spaceflight: A literature review.Clin Neurol Neurosurg. 2021 Aug;207:106755. doi: 10.1016/j.clineuro.2021.106755. Epub 2021 Jun 8. Clin Neurol Neurosurg. 2021. PMID: 34126454 Review.
-
Impacts of microgravity on amino acid metabolism during spaceflight.Exp Biol Med (Maywood). 2023 May;248(5):380-393. doi: 10.1177/15353702221139189. Epub 2023 Feb 12. Exp Biol Med (Maywood). 2023. PMID: 36775855 Free PMC article. Review.
Cited by
-
A second space age spanning omics, platforms and medicine across orbits.Nature. 2024 Aug;632(8027):995-1008. doi: 10.1038/s41586-024-07586-8. Epub 2024 Jun 11. Nature. 2024. PMID: 38862027 Free PMC article. Review.
-
Protective alleles and precision healthcare in crewed spaceflight.Nat Commun. 2024 Jul 22;15(1):6158. doi: 10.1038/s41467-024-49423-6. Nat Commun. 2024. PMID: 39039045 Free PMC article. Review.
-
Genome and clonal hematopoiesis stability contrasts with immune, cfDNA, mitochondrial, and telomere length changes during short duration spaceflight.Precis Clin Med. 2024 Apr 8;7(1):pbae007. doi: 10.1093/pcmedi/pbae007. eCollection 2024 Mar. Precis Clin Med. 2024. PMID: 38634106 Free PMC article.
-
Telomeric RNA (TERRA) increases in response to spaceflight and high-altitude climbing.Commun Biol. 2024 Jun 11;7(1):698. doi: 10.1038/s42003-024-06014-x. Commun Biol. 2024. PMID: 38862827 Free PMC article.
-
Collection of biospecimens from the inspiration4 mission establishes the standards for the space omics and medical atlas (SOMA).Nat Commun. 2024 Jun 11;15(1):4964. doi: 10.1038/s41467-024-48806-z. Nat Commun. 2024. PMID: 38862509 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

