Red2Flpe-SCON: a versatile, multicolor strategy for generating mosaic conditional knockout mice
- PMID: 38862535
- PMCID: PMC11166929
- DOI: 10.1038/s41467-024-49382-y
Red2Flpe-SCON: a versatile, multicolor strategy for generating mosaic conditional knockout mice
Abstract
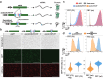
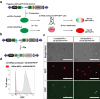
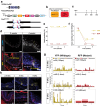
Image-based lineage tracing enables tissue turnover kinetics and lineage potentials of different adult cell populations to be investigated. Previously, we reported a genetic mouse model system, Red2Onco, which ectopically expressed mutated oncogenes together with red fluorescent proteins (RFP). This system enabled the expansion kinetics and neighboring effects of oncogenic clones to be dissected. We now report Red2Flpe-SCON: a mosaic knockout system that uses multicolor reporters to label both mutant and wild-type cells. We develop the Red2Flpe mouse line for red clone-specific Flpe expression, as well as the FRT-based SCON (Short Conditional IntrON) method to facilitate tunable conditional mosaic knockouts in mice. We use the Red2Flpe-SCON method to study Sox2 mutant clonal analysis in the esophageal epithelium of adult mice which reveal that the stem cell gene, Sox2, is less essential for adult stem cell maintenance itself, but rather for stem cell proliferation and differentiation.
© 2024. The Author(s).
Conflict of interest statement
S.W. and B.K.K. are inventors of a patent application on the SCON technology used in this study which was submitted by the Institute of Molecular Biotechnology to the European patent office (EP21172761) followed by a PCT application (WO2022/234086A1), entitled “Controlled gene expression methods and means”. The remaining authors declare no competing interests.
Figures


Similar articles
-
The transcription factor Sox2 is required for osteoblast self-renewal.Cell Death Differ. 2010 Aug;17(8):1345-53. doi: 10.1038/cdd.2010.57. Epub 2010 May 21. Cell Death Differ. 2010. PMID: 20489730 Free PMC article.
-
SCON-a Short Conditional intrON for conditional knockout with one-step zygote injection.Exp Mol Med. 2022 Dec;54(12):2188-2199. doi: 10.1038/s12276-022-00891-0. Epub 2022 Dec 9. Exp Mol Med. 2022. PMID: 36494589 Free PMC article.
-
Sox2 controls asymmetric patterning of ameloblast lineage commitment by regulation of FGF signaling in the mouse incisor.J Mol Histol. 2021 Oct;52(5):1035-1042. doi: 10.1007/s10735-021-10005-1. Epub 2021 Jul 19. J Mol Histol. 2021. PMID: 34279757
-
Transgenic mice: beyond the knockout.Am J Physiol Renal Physiol. 2011 Feb;300(2):F291-300. doi: 10.1152/ajprenal.00082.2010. Epub 2010 Nov 10. Am J Physiol Renal Physiol. 2011. PMID: 21068085 Free PMC article. Review.
-
Generating mosaics for lineage analysis in flies.Wiley Interdiscip Rev Dev Biol. 2014 Jan-Feb;3(1):69-81. doi: 10.1002/wdev.122. Epub 2013 Jun 28. Wiley Interdiscip Rev Dev Biol. 2014. PMID: 24902835 Free PMC article. Review.
Cited by
-
Redefining familial adenomatous polyposis: competition, cooperation, and the path to monoclonality.Fam Cancer. 2025 Jun 1;24(2):52. doi: 10.1007/s10689-025-00479-3. Fam Cancer. 2025. PMID: 40451939 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

