Aging and putative frailty biomarkers are altered by spaceflight
- PMID: 38862573
- PMCID: PMC11166946
- DOI: 10.1038/s41598-024-57948-5
Aging and putative frailty biomarkers are altered by spaceflight
Abstract
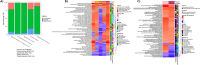
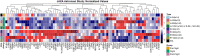
Human space exploration poses inherent risks to astronauts' health, leading to molecular changes that can significantly impact their well-being. These alterations encompass genomic instability, mitochondrial dysfunction, increased inflammation, homeostatic dysregulation, and various epigenomic changes. Remarkably, these changes bear similarities to those observed during the aging process on Earth. However, our understanding of the connection between these molecular shifts and disease development in space remains limited. Frailty syndrome, a clinical syndrome associated with biological aging, has not been comprehensively investigated during spaceflight. To bridge this knowledge gap, we leveraged murine data obtained from NASA's GeneLab, along with astronaut data gathered from the JAXA and Inspiration4 missions. Our objective was to assess the presence of biological markers and pathways related to frailty, aging, and sarcopenia within the spaceflight context. Through our analysis, we identified notable changes in gene expression patterns that may be indicative of the development of a frailty-like condition during space missions. These findings suggest that the parallels between spaceflight and the aging process may extend to encompass frailty as well. Consequently, further investigations exploring the utility of a frailty index in monitoring astronaut health appear to be warranted.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

