Klebsiella oxytoca inhibits Salmonella infection through multiple microbiota-context-dependent mechanisms
- PMID: 38862602
- PMCID: PMC11222139
- DOI: 10.1038/s41564-024-01710-0
Klebsiella oxytoca inhibits Salmonella infection through multiple microbiota-context-dependent mechanisms
Abstract
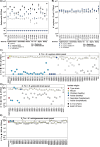
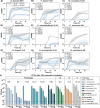
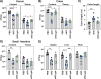
The Klebsiella oxytoca species complex is part of the human microbiome, especially during infancy and childhood. K. oxytoca species complex strains can produce enterotoxins, namely, tilimycin and tilivalline, while also contributing to colonization resistance (CR). The relationship between these seemingly contradictory roles is not well understood. Here, by coupling ex vivo assays with CRISPR-mutagenesis and various mouse models, we show that K. oxytoca provides CR against Salmonella Typhimurium. In vitro, the antimicrobial activity against various Salmonella strains depended on tilimycin production and was induced by various simple carbohydrates. In vivo, CR against Salmonella depended on toxin production in germ-free mice, while it was largely toxin-independent in mice with residual microbiota. This was linked to the relative levels of toxin-inducing carbohydrates in vivo. Finally, dulcitol utilization was essential for toxin-independent CR in gnotobiotic mice. Together, this demonstrates that nutrient availability is key to both toxin-dependent and substrate-driven competition between K. oxytoca and Salmonella.
© 2024. The Author(s).
Conflict of interest statement
L.O., M.W. and T.S. have filed a patent for the use of
Figures













References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

