Crowd-sourced benchmarking of single-sample tumor subclonal reconstruction
- PMID: 38862616
- PMCID: PMC11994449
- DOI: 10.1038/s41587-024-02250-y
Crowd-sourced benchmarking of single-sample tumor subclonal reconstruction
Abstract
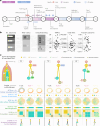
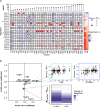
Subclonal reconstruction algorithms use bulk DNA sequencing data to quantify parameters of tumor evolution, allowing an assessment of how cancers initiate, progress and respond to selective pressures. We launched the ICGC-TCGA (International Cancer Genome Consortium-The Cancer Genome Atlas) DREAM Somatic Mutation Calling Tumor Heterogeneity and Evolution Challenge to benchmark existing subclonal reconstruction algorithms. This 7-year community effort used cloud computing to benchmark 31 subclonal reconstruction algorithms on 51 simulated tumors. Algorithms were scored on seven independent tasks, leading to 12,061 total runs. Algorithm choice influenced performance substantially more than tumor features but purity-adjusted read depth, copy-number state and read mappability were associated with the performance of most algorithms on most tasks. No single algorithm was a top performer for all seven tasks and existing ensemble strategies were unable to outperform the best individual methods, highlighting a key research need. All containerized methods, evaluation code and datasets are available to support further assessment of the determinants of subclonal reconstruction accuracy and development of improved methods to understand tumor evolution.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: I.L. is a consultant for PACT Pharma, Inc. and is an equity holder, board member and consultant for ennov1, LLC. P.C.B. sits on the scientific advisory boards of BioSymetrics, Inc. and Intersect Diagnostics, Inc. and previously sat on that of Sage Bionetworks. A.S. is a shareholder of Illumina, Inc.
Figures












References
-
- Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell144, 646–674 (2011). - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

