Altered salivary microbiota associated with high-sugar beverage consumption
- PMID: 38862651
- PMCID: PMC11167035
- DOI: 10.1038/s41598-024-64324-w
Altered salivary microbiota associated with high-sugar beverage consumption
Abstract
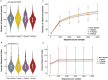
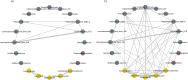
The human oral microbiome may alter oral and systemic disease risk. Consuming high sugar content beverages (HSB) can lead to caries development by altering the microbial composition in dental plaque, but little is known regarding HSB-specific oral microbial alterations. Therefore, we conducted a large, population-based study to examine associations of HSB intake with oral microbiome diversity and composition. Using mouthwash samples of 989 individuals in two nationwide U.S. cohorts, bacterial 16S rRNA genes were amplified, sequenced, and assigned to bacterial taxa. HSB intake was quantified from food frequency questionnaires as low (< 1 serving/week), medium (1-3 servings/week), or high (> 3 servings/week). We assessed overall bacterial diversity and presence of specific taxa with respect to HSB intake in each cohort separately and combined in a meta-analysis. Consistently in the two cohorts, we found lower species richness in high HSB consumers (> 3 cans/week) (p = 0.027), and that overall bacterial community profiles differed from those of non-consumers (PERMANOVA p = 0.040). Specifically, presence of a network of commensal bacteria (Lachnospiraceae, Peptostreptococcaceae, and Alloprevotella rava) was less common in high compared to non-consumers, as were other species including Campylobacter showae, Prevotella oulorum, and Mycoplasma faucium. Presence of acidogenic bacteria Bifodobacteriaceae and Lactobacillus rhamnosus was more common in high consumers. Abundance of Fusobacteriales and its genus Leptotrichia, Lachnoanaerobaculum sp., and Campylobacter were lower with higher HSB consumption, and their abundances were correlated. No significant interaction was found for these associations with diabetic status or with microbial markers for caries (S. mutans) and periodontitis (P. gingivalis). Our results suggest that soft drink intake may alter the salivary microbiota, with consistent results across two independent cohorts. The observed perturbations of overrepresented acidogenic bacteria and underrepresented commensal bacteria in high HSB consumers may have implications for oral and systemic disease risk.
Keywords: High-sugar beverage consumption; Oral health; Oral microbiome; Population-based study; Salivary microbiota; Soda.
© 2024. The Author(s).
Conflict of interest statement
XF: Director, Real-World Evidence, Data Science (Jazz Pharmaceuticals). All other authors do not hold any competing interest.
Figures

Similar articles
-
The influence of diet, saliva, and dental history on the oral microbiome in healthy, caries-free Australian adults.Sci Rep. 2025 May 28;15(1):18755. doi: 10.1038/s41598-025-03455-0. Sci Rep. 2025. PMID: 40436959 Free PMC article.
-
Salivary microbiota characterization of Yerba Mate consumers in Uruguay.Clin Oral Investig. 2025 Feb 15;29(2):131. doi: 10.1007/s00784-025-06209-4. Clin Oral Investig. 2025. PMID: 39954023
-
Identification of Salivary Microbiota and Its Association With Host Inflammatory Mediators in Periodontitis.Front Cell Infect Microbiol. 2019 Jun 21;9:216. doi: 10.3389/fcimb.2019.00216. eCollection 2019. Front Cell Infect Microbiol. 2019. PMID: 31281801 Free PMC article.
-
The oral microbiome and human health.J Oral Sci. 2017;59(2):201-206. doi: 10.2334/josnusd.16-0856. J Oral Sci. 2017. PMID: 28637979 Review.
-
Environmental interventions to reduce the consumption of sugar-sweetened beverages and their effects on health.Cochrane Database Syst Rev. 2019 Jun 12;6(6):CD012292. doi: 10.1002/14651858.CD012292.pub2. Cochrane Database Syst Rev. 2019. PMID: 31194900 Free PMC article.
Cited by
-
The influence of diet, saliva, and dental history on the oral microbiome in healthy, caries-free Australian adults.Sci Rep. 2025 May 28;15(1):18755. doi: 10.1038/s41598-025-03455-0. Sci Rep. 2025. PMID: 40436959 Free PMC article.
-
Oral microbiota in preschoolers with rampant caries: a matched case-control study.Appl Microbiol Biotechnol. 2024 Dec 11;108(1):533. doi: 10.1007/s00253-024-13362-5. Appl Microbiol Biotechnol. 2024. PMID: 39661115 Free PMC article.
-
Impact of sugar-sweetened beverages on salivary parameters: A systematic review & meta-analysis.Evid Based Dent. 2025 May 9. doi: 10.1038/s41432-025-01147-2. Online ahead of print. Evid Based Dent. 2025. PMID: 40346271
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

