Construction of a nomogram for predicting HNSCC distant metastasis and identification of EIF5A as a hub gene
- PMID: 38862693
- PMCID: PMC11166653
- DOI: 10.1038/s41598-024-64197-z
Construction of a nomogram for predicting HNSCC distant metastasis and identification of EIF5A as a hub gene
Abstract
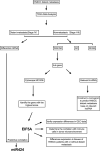
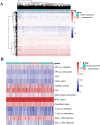
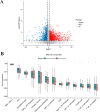
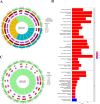
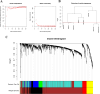
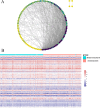
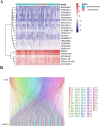
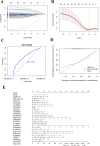
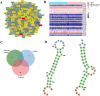
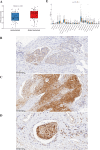
Patients with distant metastasis of head and neck squamous cell carcinoma (HNSCC) often have a poor prognosis. However, early diagnosis of distant metastasis is challenging in clinical practice, and distant metastasis is often only detected in the late stages of tumor metastasis through imaging techniques. In this study, we utilized data from HNSCC patients collected from the TCGA database. Patients were divided into distant metastasis and nonmetastasis groups based on the tumor-node-metastasis (TNM) stage. We analyzed the differentially expressed genes between the two groups (DM/non-M DEGs) and their associated lncRNAs and generated a predictive model based on 23 lncRNAs that were significantly associated with the occurrence of distant metastasis in HNSCC patients. On this basis, we built a nomogram to predict the distant metastasis of HNSCC patients. Moreover, through WGCNA and Cytoscape software analysis of DM/non-M DEGs, we identified the gene most closely related to HNSCC distant metastasis: EIF5A. Our findings were validated using GEO data; EIF5A expression was significantly increased in the tumor tissues of HNSCC patients with distant metastasis. We then predicted miRNAs that can directly bind to EIF5A via the TargetScan and miRWalk websites, intersected them with differentially expressed miRNAs in the two groups from the TCGA cohort, and identified the only overlapping miRNA, miR-424; we predicted the direct binding site of EIF5A and miR-424 via the miRWalk website. Immunohistochemistry further revealed high expression of EIF5A in the primary tumor tissue of HNSCC patients with distant metastasis. These results provide a new perspective for the early diagnosis of distant metastasis in HNSCC patients and the study of the mechanisms underlying HNSCC distant metastasis.
Keywords: Distant metastasis; EIF5A; HNSCC; Nomogram; miR-424.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Excavating novel diagnostic and prognostic long non-coding RNAs (lncRNAs) for head and neck squamous cell carcinoma: an integrated bioinformatics analysis of competing endogenous RNAs (ceRNAs) and gene co-expression networks.Bioengineered. 2021 Dec;12(2):12821-12838. doi: 10.1080/21655979.2021.2003925. Bioengineered. 2021. PMID: 34898376 Free PMC article.
-
Construction of an 11-microRNA-based signature and a prognostic nomogram to predict the overall survival of head and neck squamous cell carcinoma patients.BMC Genomics. 2020 Oct 6;21(1):691. doi: 10.1186/s12864-020-07104-w. BMC Genomics. 2020. PMID: 33023466 Free PMC article.
-
Transcriptome analysis reveals the link between lncRNA-mRNA co-expression network and tumor immune microenvironment and overall survival in head and neck squamous cell carcinoma.BMC Med Genomics. 2020 Mar 30;13(1):57. doi: 10.1186/s12920-020-0707-0. BMC Med Genomics. 2020. PMID: 32228580 Free PMC article.
-
Long noncoding RNA MYOSLID promotes invasion and metastasis by modulating the partial epithelial-mesenchymal transition program in head and neck squamous cell carcinoma.J Exp Clin Cancer Res. 2019 Jun 25;38(1):278. doi: 10.1186/s13046-019-1254-4. J Exp Clin Cancer Res. 2019. PMID: 31238980 Free PMC article.
-
Insights into metastatic roadmap of head and neck cancer squamous cell carcinoma based on clinical, histopathological and molecular profiles.Mol Biol Rep. 2024 Apr 29;51(1):597. doi: 10.1007/s11033-024-09476-8. Mol Biol Rep. 2024. PMID: 38683372 Free PMC article. Review.
References
-
- Kunieda F, et al. Randomized phase II/III trial of post-operative chemoradiotherapy comparing 3-weekly cisplatin with weekly cisplatin in high-risk patients with squamous cell carcinoma of head and neck: Japan Clinical Oncology Group Study (JCOG1008) Jpn. J. Clin. Oncol. 2014;44:770–774. doi: 10.1093/jjco/hyu067. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

