Pharmacological inhibition of RUNX1 reduces infarct size after acute myocardial infarction in rats and underlying mechanism revealed by proteomics implicates repressed cathepsin levels
- PMID: 38862712
- PMCID: PMC11166773
- DOI: 10.1007/s10142-024-01391-2
Pharmacological inhibition of RUNX1 reduces infarct size after acute myocardial infarction in rats and underlying mechanism revealed by proteomics implicates repressed cathepsin levels
Abstract
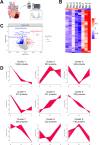
Myocardial infarction (MI) results in prolonged ischemia and the subsequent cell death leads to heart failure which is linked to increased deaths or hospitalizations. New therapeutic targets are urgently needed to prevent cell death and reduce infarct size among patients with MI. Runt-related transcription factor-1 (RUNX1) is a master-regulator transcription factor intensively studied in the hematopoietic field. Recent evidence showed that RUNX1 has a critical role in cardiomyocytes post-MI. The increased RUNX1 expression in the border zone of the infarct heart contributes to decreased cardiac contractile function and can be therapeutically targeted to protect against adverse cardiac remodelling. This study sought to investigate whether pharmacological inhibition of RUNX1 function has an impact on infarct size following MI. In this work we demonstrate that inhibiting RUNX1 with a small molecule inhibitor (Ro5-3335) reduces infarct size in an in vivo rat model of acute MI. Proteomics study using data-independent acquisition method identified increased cathepsin levels in the border zone myocardium following MI, whereas heart samples treated by RUNX1 inhibitor present decreased cathepsin levels. Cathepsins are lysosomal proteases which have been shown to orchestrate multiple cell death pathways. Our data illustrate that inhibition of RUNX1 leads to reduced infarct size which is associated with the suppression of cathepsin expression. This study demonstrates that pharmacologically antagonizing RUNX1 reduces infarct size in a rat model of acute MI and unveils a link between RUNX1 and cathepsin-mediated cell death, suggesting that RUNX1 is a novel therapeutic target that could be exploited clinically to limit infarct size after an acute MI.
Keywords: Cardiac protection; Cathepsin; Myocardial infarction; Runx1; Therapeutic target.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Ribonucleicacid interference or small molecule inhibition of Runx1 in the border zone prevents cardiac contractile dysfunction following myocardial infarction.Cardiovasc Res. 2023 Dec 19;119(16):2663-2671. doi: 10.1093/cvr/cvad107. Cardiovasc Res. 2023. PMID: 37433039 Free PMC article.
-
Dihydrolycorine Attenuates Cardiac Fibrosis and Dysfunction by Downregulating Runx1 following Myocardial Infarction.Oxid Med Cell Longev. 2021 Oct 23;2021:8528239. doi: 10.1155/2021/8528239. eCollection 2021. Oxid Med Cell Longev. 2021. PMID: 34725565 Free PMC article.
-
Runx1 Deficiency Protects Against Adverse Cardiac Remodeling After Myocardial Infarction.Circulation. 2018 Jan 2;137(1):57-70. doi: 10.1161/CIRCULATIONAHA.117.028911. Epub 2017 Oct 13. Circulation. 2018. PMID: 29030345 Free PMC article.
-
RUNX1: an emerging therapeutic target for cardiovascular disease.Cardiovasc Res. 2020 Jul 1;116(8):1410-1423. doi: 10.1093/cvr/cvaa034. Cardiovasc Res. 2020. PMID: 32154891 Free PMC article. Review.
-
Angiogenesis after acute myocardial infarction.Cardiovasc Res. 2021 Apr 23;117(5):1257-1273. doi: 10.1093/cvr/cvaa287. Cardiovasc Res. 2021. PMID: 33063086 Review.
Cited by
-
Acid-responsive aggregated carrot-derived nanoantioxidants alleviate oxidative stress and restore osteoblast activity.J Nanobiotechnology. 2025 Mar 12;23(1):206. doi: 10.1186/s12951-025-03235-y. J Nanobiotechnology. 2025. PMID: 40075427 Free PMC article.
References
-
- Burgener SS, Leborgne NGF, Snipas SJ, Salvesen GS, Bird PI, Benarafa C. Cathepsin G Inhibition by Serpinb1 and Serpinb6 Prevents Programmed Necrosis in Neutrophils and Monocytes and Reduces GSDMD-Driven Inflammation. Cell Rep. 2019;27(12):3646–3656.e3645. doi: 10.1016/j.celrep.2019.05.065. - DOI - PMC - PubMed
-
- Campden RI, Warren AL, Greene CJ, Chiriboga JA, Arnold CR, Aggarwal D, McKenna N, Sandall CF, MacDonald JA, Yates RM. Extracellular cathepsin Z signals through the α(5) integrin and augments NLRP3 inflammasome activation. J Biol Chem. 2022;298(1):101459. doi: 10.1016/j.jbc.2021.101459. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

