IL-12 induces a B cell-intrinsic IL-12/IFNγ feed-forward loop promoting extrafollicular B cell responses
- PMID: 38862796
- PMCID: PMC11992614
- DOI: 10.1038/s41590-024-01858-1
IL-12 induces a B cell-intrinsic IL-12/IFNγ feed-forward loop promoting extrafollicular B cell responses
Abstract
While some infections elicit germinal centers, others produce only extrafollicular responses. The mechanisms controlling these dichotomous fates are poorly understood. We identify IL-12 as a cytokine switch, acting directly on B cells to promote extrafollicular and suppress germinal center responses. IL-12 initiates a B cell-intrinsic feed-forward loop between IL-12 and IFNγ, amplifying IFNγ production, which promotes proliferation and plasmablast differentiation from mouse and human B cells, in synergy with IL-12. IL-12 sustains the expression of a portion of IFNγ-inducible genes. Together, they also induce unique gene changes, reflecting both IFNγ amplification and cooperative effects between both cytokines. In vivo, cells lacking both IL-12 and IFNγ receptors are more impaired in plasmablast production than those lacking either receptor alone. Further, B cell-derived IL-12 enhances both plasmablast responses and T helper 1 cell commitment. Thus, B cell-derived IL-12, acting on T and B cells, determines the immune response mode, with implications for vaccines, pathogen protection and autoimmunity.
© 2024. The Author(s), under exclusive licence to Springer Nature America, Inc.
Conflict of interest statement
Competing Interests Statement
The authors declare no competing interests.
Figures










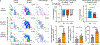



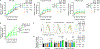


References
-
- Weisel F & Shlomchik M Memory B Cells of Mice and Humans. Annu Rev Immunol 35, 255–284 (2017), PMID28142324 - PubMed
-
- Liu X, Yao J, Zhao Y, Wang J & Qi H Heterogeneous plasma cells and long-lived subsets in response to immunization, autoantigen and microbiota. Nat Immunol 23, 1564–1576 (2022), PMID36316480 - PubMed
-
- William J, Euler C, Christensen S & Shlomchik MJ Evolution of autoantibody responses via somatic hypermutation outside of germinal centers. Science 297, 2066–2070 (2002), PMID12242446 - PubMed
Methods-only References
-
- Sonoda E et al. B cell development under the condition of allelic inclusion. Immunity 6, 225–233 (1997), PMID9075923 - PubMed
-
- Hoiseth SK & Stocker BA Aromatic-dependent Salmonella typhimurium are non-virulent and effective as live vaccines. Nature 291, 238–239 (1981), PMID7015147 - PubMed
-
- Liao Y, Smyth GK & Shi W featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014), PMID24227677 - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

