Transcriptome analysis of the Japanese eel (Anguilla japonica) during larval metamorphosis
- PMID: 38862878
- PMCID: PMC11165803
- DOI: 10.1186/s12864-024-10459-z
Transcriptome analysis of the Japanese eel (Anguilla japonica) during larval metamorphosis
Abstract
Background: Anguillid eels spend their larval period as leptocephalus larvae that have a unique and specialized body form with leaf-like and transparent features, and they undergo drastic metamorphosis to juvenile glass eels. Less is known about the transition of leptocephali to the glass eel stage, because it is difficult to catch the metamorphosing larvae in the open ocean. However, recent advances in rearing techniques for the Japanese eel have made it possible to study the larval metamorphosis of anguillid eels. In the present study, we investigated the dynamics of gene expression during the metamorphosis of Japanese eel leptocephali using RNA sequencing.
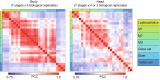
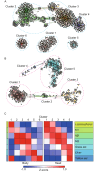
Results: During metamorphosis, Japanese eels were classified into 7 developmental stages according to their morphological characteristics, and RNA sequencing was used to collect gene expression data from each stage. A total of 354.8 million clean reads were generated from the body and 365.5 million from the head, after the processing of raw reads. For filtering of genes that characterize developmental stages, a classification model created by a Random Forest algorithm was built. Using the importance of explanatory variables feature obtained from the created model, we identified 46 genes selected in the body and 169 genes selected in the head that were defined as the "most characteristic genes" during eel metamorphosis. Next, network analysis and subsequently gene clustering were conducted using the most characteristic genes and their correlated genes, and then 6 clusters in the body and 5 clusters in the head were constructed. Then, the characteristics of the clusters were revealed by Gene Ontology (GO) enrichment analysis. The expression patterns and GO terms of each stage were consistent with previous observations and experiments during the larval metamorphosis of the Japanese eel.
Conclusion: Genome and transcriptome resources have been generated for metamorphosing Japanese eels. Genes that characterized metamorphosis of the Japanese eel were identified through statistical modeling by a Random Forest algorithm. The functions of these genes were consistent with previous observations and experiments during the metamorphosis of anguillid eels.
Keywords: Anguilla japonica; Eel genome; Leptocephali; Metamorphosis; Transcriptome.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Developmental features of Japanese eels, Anguilla japonica, from the late leptocephalus to the yellow eel stages: an early metamorphosis to the eel-like form and a prolonged transition to the juvenile.J Fish Biol. 2022 Feb;100(2):454-473. doi: 10.1111/jfb.14956. Epub 2021 Dec 28. J Fish Biol. 2022. PMID: 34813089
-
Location, size and age at onset of metamorphosis in the Japanese eel Anguilla japonica.J Fish Biol. 2018 May;92(5):1342-1358. doi: 10.1111/jfb.13590. Epub 2018 Mar 14. J Fish Biol. 2018. PMID: 29537077
-
Characterization of triglycerides during early development of the Japanese eel (Anguilla japonica).Comp Biochem Physiol A Mol Integr Physiol. 2022 Mar;265:111125. doi: 10.1016/j.cbpa.2021.111125. Epub 2021 Dec 11. Comp Biochem Physiol A Mol Integr Physiol. 2022. PMID: 34906629
-
A century of research on the larval distributions of the Atlantic eels: a re-examination of the data.Biol Rev Camb Philos Soc. 2015 Nov;90(4):1035-64. doi: 10.1111/brv.12144. Epub 2014 Oct 8. Biol Rev Camb Philos Soc. 2015. PMID: 25291986 Review.
-
Migratory life cycle of Anguilla anguilla: a mirror symmetry with A. japonica.J Fish Biol. 2025 Feb;106(2):138-156. doi: 10.1111/jfb.15966. Epub 2024 Oct 23. J Fish Biol. 2025. PMID: 39439402 Review.
References
-
- Kyle HM. The asymmetry, metamorphosis and origin of flat-fishes. Philos Trans R Soc Lond B. 1923;211:75–129. doi: 10.1098/rstb.1923.0002. - DOI
-
- Policansky D. The asymmetry of flounders. Sci Am. 1982;246:96–102. doi: 10.1038/scientificamerican0582-116. - DOI
-
- Ahlstrom EH, Amaoka DA, Hensley DA, Moser HG, Sumida BY. Pleuronectiformes: development, ontogeny and systematics of fishes. Am Soc Ichthyol Herpetol. 1984;1:640–70.
-
- Miwa S, Inui Y. Histological changes in the pituitary-thyroid axis during spontaneous and artificially-induced metamorphosis of larvae of the flounder Paralichtys Olivaceus. Cell Tissue Res. 1987;249:117–23. doi: 10.1007/BF00215425. - DOI
MeSH terms
LinkOut - more resources
Full Text Sources

