Genome-wide identification, molecular evolution and expression analysis of the B-box gene family in mung bean (Vigna radiata L.)
- PMID: 38862892
- PMCID: PMC11167828
- DOI: 10.1186/s12870-024-05236-9
Genome-wide identification, molecular evolution and expression analysis of the B-box gene family in mung bean (Vigna radiata L.)
Abstract
Background: Mung bean (Vigna radiata L.) is an important warm-season grain legume. Adaptation to extreme environmental conditions, supported by evolution, makes mung bean a rich gene pool for stress tolerance traits. The exploration of resistance genes will provide important genetic resources and a theoretical basis for strengthening mung bean breeding. B-box (BBX) proteins play a major role in developmental processes and stress responses. However, the identification and analysis of the mung bean BBX gene family are still lacking.
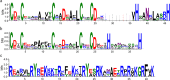
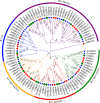
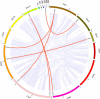
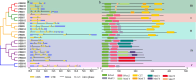
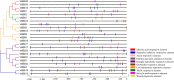
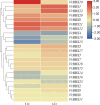
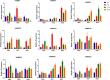
Results: In this study, 23 VrBBX genes were identified through comprehensive bioinformatics analysis and named based on their physical locations on chromosomes. All the VrBBXs were divided into five groups based on their phylogenetic relationships, the number of B-box they contained and whether there was an additional CONSTANS, CO-like and TOC1 (CCT) domain. Homology and collinearity analysis indicated that the BBX genes in mung bean and other species had undergone a relatively conservative evolution. Gene duplication analysis showed that only chromosomal segmental duplication contributed to the expansion of VrBBX genes and that most of the duplicated gene pairs experienced purifying selection pressure during evolution. Gene structure and motif analysis revealed that VrBBX genes clustered in the same group shared similar structural characteristics. An analysis of cis-acting elements indicated that elements related to stress and hormone responses were prevalent in the promoters of most VrBBXs. The RNA-seq data analysis and qRT-PCR of nine VrBBX genes demonstrated that VrBBX genes may play a role in response to environmental stress. Moreover, VrBBX5, VrBBX10 and VrBBX12 are important candidate genes for plant stress response.
Conclusions: In this study, we systematically analyzed the genomic characteristics and expression patterns of the BBX gene family under ABA, PEG and NaCl treatments. The results will help us better understand the complexity of the BBX gene family and provide valuable information for future functional characteristics of specific genes in this family.
Keywords: B-box gene family; Abiotic stresses; Evolution; Expression profiles; Mung bean.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Genome-wide analysis of OSCA gene family members in Vigna radiata and their involvement in the osmotic response.BMC Plant Biol. 2021 Sep 7;21(1):408. doi: 10.1186/s12870-021-03184-2. BMC Plant Biol. 2021. PMID: 34493199 Free PMC article.
-
Genome-wide identification and expression profiles of AP2/ERF transcription factor family in mung bean (Vigna radiata L.).J Appl Genet. 2022 May;63(2):223-236. doi: 10.1007/s13353-021-00675-8. Epub 2022 Jan 6. J Appl Genet. 2022. PMID: 34989979
-
Genome-wide analyses of the mung bean NAC gene family reveals orthologs, co-expression networking and expression profiling under abiotic and biotic stresses.BMC Plant Biol. 2022 Jul 15;22(1):343. doi: 10.1186/s12870-022-03716-4. BMC Plant Biol. 2022. PMID: 35836131 Free PMC article.
-
Genome-wide identification and expression analysis of the Dof gene family reveals their involvement in hormone response and abiotic stresses in sunflower (Helianthus annuus L.).Gene. 2024 Jun 5;910:148336. doi: 10.1016/j.gene.2024.148336. Epub 2024 Mar 4. Gene. 2024. PMID: 38447680 Review.
-
Mung Bean (Vigna radiata L.): Bioactive Polyphenols, Polysaccharides, Peptides, and Health Benefits.Nutrients. 2019 May 31;11(6):1238. doi: 10.3390/nu11061238. Nutrients. 2019. PMID: 31159173 Free PMC article. Review.
Cited by
-
Genome-wide identification and expression analysis of phytochrome gene family in Aikang58 wheat (Triticum aestivum L.).Front Plant Sci. 2025 Jan 21;15:1520457. doi: 10.3389/fpls.2024.1520457. eCollection 2024. Front Plant Sci. 2025. PMID: 39906238 Free PMC article.
-
Morpho-biochemical and molecular profiling for Charcoal Rot (CR) disease resistance in Mung Bean [Vigna radiata (L.) Wilczek] landraces.Mol Biol Rep. 2024 Dec 24;52(1):76. doi: 10.1007/s11033-024-10187-3. Mol Biol Rep. 2024. PMID: 39718653
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

