Whole-genome SNP allele frequency differences between Tibetan and Large white pigs reveal genes associated with skeletal muscle growth
- PMID: 38862895
- PMCID: PMC11167949
- DOI: 10.1186/s12864-024-10508-7
Whole-genome SNP allele frequency differences between Tibetan and Large white pigs reveal genes associated with skeletal muscle growth
Abstract
Background: The skeletal muscle growth rate and body size of Tibetan pigs (TIB) are lower than Large white pigs (LW). However, the underlying genetic basis attributing to these differences remains uncertain. To address this knowledge gap, the present study employed whole-genome sequencing of TIB (slow growth) and LW (fast growth) individuals, and integrated with existing NCBI sequencing datasets of TIB and LW individuals, enabling the identification of a comprehensive set of genetic variations for each breed. The specific and predominant SNPs in the TIB and LW populations were detected by using a cutoff value of 0.50 for SNP allele frequency and absolute allele frequency differences (△AF) between the TIB and LW populations.
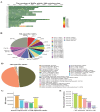
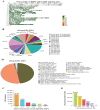
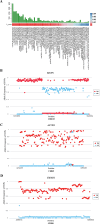
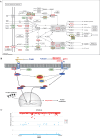
Results: A total of 21,767,938 SNPs were retrieved from 44 TIB and 29 LW genomes. The analysis detected 2,893,106 (13.29%) and 813,310 (3.74%) specific and predominant SNPs in the TIB and LW populations, and annotated to 24,560 genes. Further GO analysis revealed 291 genes involved in biological processes related to striated and/or skeletal muscle differentiation, proliferation, hypertrophy, regulation of striated muscle cell differentiation and proliferation, and myoblast differentiation and fusion. These 291 genes included crucial regulators of muscle cell determination, proliferation, differentiation, and hypertrophy, such as members of the Myogenic regulatory factors (MRF) (MYOD, MYF5, MYOG, MYF6) and Myocyte enhancer factor 2 (MEF2) (MEF2A, MEF2C, MEF2D) families, as well as muscle growth inhibitors (MSTN, ACVR1, and SMAD1); KEGG pathway analysis revealed 106 and 20 genes were found in muscle growth related positive and negative regulatory signaling pathways. Notably, genes critical for protein synthesis, such as MTOR, IGF1, IGF1R, IRS1, INSR, and RPS6KA6, were implicated in these pathways.
Conclusion: This study employed an effective methodology to rigorously identify the potential genes associated with skeletal muscle development. A substantial number of SNPs and genes that potentially play roles in the divergence observed in skeletal muscle growth between the TIB and LW breeds were identified. These findings offer valuable insights into the genetic underpinnings of skeletal muscle development and present opportunities for enhancing meat production through pig breeding.
Keywords: Large white pigs; Predominant SNPs; Skeletal muscle growth; Specific SNPs; Tibetan pigs; Whole-genome SNPs.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Investigation of single nucleotide polymorphisms in differentially expressed genes and proteins reveals the genetic basis of skeletal muscle growth differences between Tibetan and Large White pigs.Anim Biosci. 2024 Dec;37(12):2021-2032. doi: 10.5713/ab.24.0135. Epub 2024 Jun 25. Anim Biosci. 2024. PMID: 38938033 Free PMC article.
-
Comparative analyses by sequencing of transcriptomes during skeletal muscle development between pig breeds differing in muscle growth rate and fatness.PLoS One. 2011;6(5):e19774. doi: 10.1371/journal.pone.0019774. Epub 2011 May 26. PLoS One. 2011. PMID: 21637832 Free PMC article.
-
Whole genome and transcriptome analyses identify genetic markers associated with growth traits in Qinchuan black pig.BMC Genomics. 2025 May 12;26(1):469. doi: 10.1186/s12864-025-11627-5. BMC Genomics. 2025. PMID: 40355827 Free PMC article.
-
MicroRNA in Skeletal Muscle: Its Crucial Roles in Signal Proteins, Mus cle Fiber Type, and Muscle Protein Synthesis.Curr Protein Pept Sci. 2017;18(6):579-588. doi: 10.2174/1389203717666160621122405. Curr Protein Pept Sci. 2017. PMID: 27341987 Review.
-
Epigenetic regulation of skeletal muscle development and differentiation.Subcell Biochem. 2013;61:139-50. doi: 10.1007/978-94-007-4525-4_7. Subcell Biochem. 2013. PMID: 23150250 Review.
Cited by
-
miRNA-34a-5p inhibits chicken myoblasts proliferation and differentiation via NOTCH1 inhibition.Poult Sci. 2025 Mar;104(3):104895. doi: 10.1016/j.psj.2025.104895. Epub 2025 Feb 7. Poult Sci. 2025. PMID: 39970517 Free PMC article.
References
-
- Zhu L, Li MZ, Li XW, Shuai SR, Liu HF, Wang JR, et al. Distinct expression patterns of genes Associated with muscle growth and adipose deposition in Tibetan pigs: a possible adaptive mechanism for high Altitude conditions. High Alt Med Biol. 2009;10(1):45–55. doi: 10.1089/ham.2008.1042. - DOI - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

