Single molecule array measures of LRRK2 kinase activity in serum link Parkinson's disease severity to peripheral inflammation
- PMID: 38862989
- PMCID: PMC11167795
- DOI: 10.1186/s13024-024-00738-4
Single molecule array measures of LRRK2 kinase activity in serum link Parkinson's disease severity to peripheral inflammation
Abstract
Background: LRRK2-targeting therapeutics that inhibit LRRK2 kinase activity have advanced to clinical trials in idiopathic Parkinson's disease (iPD). LRRK2 phosphorylates Rab10 on endolysosomes in phagocytic cells to promote some types of immunological responses. The identification of factors that regulate LRRK2-mediated Rab10 phosphorylation in iPD, and whether phosphorylated-Rab10 levels change in different disease states, or with disease progression, may provide insights into the role of Rab10 phosphorylation in iPD and help guide therapeutic strategies targeting this pathway.
Methods: Capitalizing on past work demonstrating LRRK2 and phosphorylated-Rab10 interact on vesicles that can shed into biofluids, we developed and validated a high-throughput single-molecule array assay to measure extracellular pT73-Rab10. Ratios of pT73-Rab10 to total Rab10 measured in biobanked serum samples were compared between informative groups of transgenic mice, rats, and a deeply phenotyped cohort of iPD cases and controls. Multivariable and weighted correlation network analyses were used to identify genetic, transcriptomic, clinical, and demographic variables that predict the extracellular pT73-Rab10 to total Rab10 ratio.
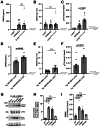
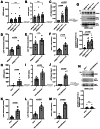
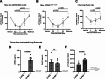
Results: pT73-Rab10 is absent in serum from Lrrk2 knockout mice but elevated by LRRK2 and VPS35 mutations, as well as SNCA expression. Bone-marrow transplantation experiments in mice show that serum pT73-Rab10 levels derive primarily from circulating immune cells. The extracellular ratio of pT73-Rab10 to total Rab10 is dynamic, increasing with inflammation and rapidly decreasing with LRRK2 kinase inhibition. The ratio of pT73-Rab10 to total Rab10 is elevated in iPD patients with greater motor dysfunction, irrespective of disease duration, age, sex, or the usage of PD-related or anti-inflammatory medications. pT73-Rab10 to total Rab10 ratios are associated with neutrophil degranulation, antigenic responses, and suppressed platelet activation.
Conclusions: The extracellular serum ratio of pT73-Rab10 to total Rab10 is a novel pharmacodynamic biomarker for LRRK2-linked innate immune activation associated with disease severity in iPD. We propose that those iPD patients with higher serum pT73-Rab10 levels may benefit from LRRK2-targeting therapeutics that mitigate associated deleterious immunological responses.
Keywords: Blood-based biomarkers; Inflammation; Neurodegeneration; Single-molecule array assays; Transcriptomic analysis.
© 2024. The Author(s).
Conflict of interest statement
A.B.W. has served as a member of The Michael J. Fox Executive Foundation (MJFF) Scientific Advisory Board and is a paid consultant for SciNeuro, Inc, and has been a paid consultant for EscapeBio Inc.; and has received research grants from Biogen Inc. and EscapeBio, Inc., as well as MJFF, ASAP Foundation, Parkinson’s Foundation, and the National Institutes of Health (NIH). A.B.W. is part owner of a series of LRRK2 kinase inhibitors (WO 2013166276) and part owner of induced-pluripotent stem cell lines of early-onset PD distributed by Cedars Sinai. L.H.S. is on the Scientific Advisory Board of Lucy Therapeutics. D.G.S. is a member of the faculty of the University of Alabama at Birmingham and is supported by endowment and University funds. Dr. Standaert is an investigator in studies funded by Abbvie, Inc., the American Parkinson Disease Association, the Michael J. Fox Foundation for Parkinson Research, The National Parkinson Foundation, Alabama Department of Commerce, Alabama Innovation Fund, Genetech, the Department of Defense, and the NIH. He has a clinical practice and is compensated for these activities through the University of Alabama Health Services Foundation. He serves as Deputy Editor for the journal Movement Disorders and is compensated for this role by the International Parkinson and Movement Disorders Society. He has served as a consultant for or received honoraria from Abbvie Inc., Alnylam Pharmaceutics, Appello, Biohaven Pharmaceuticals, Inc., BlueRock Therapeutics, Coave Therapeutics Curium Pharma, F. Hoffman-La Roche, Eli Lilly USA, Sanofi-Aventis, and Theravance, Inc. He has also received book royalties from McGraw-Hill Publishers. T.A.Y. has active grants from the American Parkinson Disease Association, Travere Therapeutics, and the NIH (RF1NS115767-A1, R01NS112203, P50NS108675, U01NS104326, R13GM109532, T32GM008361). T.A.Y. serves on the Scientific Advisory Board for Parkinson’s Foundation. TAY has received honorarium for presentations and panels from the Movement Disorders Society and for grant reviews from NIH. She has a U.S. Patent #7,919,262 on the use of 14-3-3s in neurodegeneration.
Figures





Update of
-
Single molecule array measures of LRRK2 kinase activity in serum link Parkinson's disease severity to peripheral inflammation.bioRxiv [Preprint]. 2024 Apr 17:2024.04.15.589570. doi: 10.1101/2024.04.15.589570. bioRxiv. 2024. Update in: Mol Neurodegener. 2024 Jun 11;19(1):47. doi: 10.1186/s13024-024-00738-4. PMID: 38659797 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

