Gene silencing in the aedine cell lines C6/36 and U4.4 using long double-stranded RNA
- PMID: 38863029
- PMCID: PMC11167938
- DOI: 10.1186/s13071-024-06340-3
Gene silencing in the aedine cell lines C6/36 and U4.4 using long double-stranded RNA
Abstract
Background: RNA interference (RNAi) is a target-specific gene silencing method that can be used to determine gene functions and investigate host-pathogen interactions, as well as facilitating the development of ecofriendly pesticides. Commercially available transfection reagents (TRs) can improve the efficacy of RNAi. However, we currently lack a product and protocol for the transfection of insect cell lines with long double-stranded RNA (dsRNA).
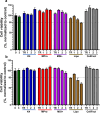
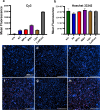
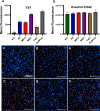
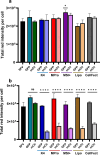
Methods: We used agarose gel electrophoresis to determine the capacity of eight TRs to form complexes with long dsRNA. A CellTiter-Glo assay was then used to assess the cytotoxicity of the resulting lipoplexes. We also measured the cellular uptake of dsRNA by fluorescence microscopy using the fluorophore Cy3 as a label. Finally, we analyzed the TRs based on their transfection efficacy and compared the RNAi responses of Aedes albopictus C6/36 and U4.4 cells by knocking down an mCherry reporter Semliki Forest virus in both cell lines.
Results: The TRs from Biontex (K4, Metafectene Pro, and Metafectene SI+) showed the best complexing capacity and the lowest dsRNA:TR ratio needed for complete complex formation. Only HiPerFect was unable to complex the dsRNA completely, even at a ratio of 1:9. Most of the complexes containing mCherry-dsRNA were nontoxic at 2 ng/µL, but Lipofectamine 2000 was toxic at 1 ng/µL in U4.4 cells and at 2 ng/µL in C6/36 cells. The transfection of U4.4 cells with mCherry-dsRNA/TR complexes achieved significant knockdown of the virus reporter. Comparison of the RNAi response in C6/36 and U4.4 cells suggested that C6/36 cells lack the antiviral RNAi response because there was no significant knockdown of the virus reporter in any of the treatments.
Conclusions: C6/36 cells have an impaired RNAi response as previously reported. This investigation provides valuable information for future RNAi experiments by showing how to mitigate the adverse effects attributed to TRs. This will facilitate the judicious selection of TRs and transfection conditions conducive to RNAi research in mosquitoes.
Keywords: Aedes albopictus; Arbovirus; Cell culture; RNAi; Semliki Forest virus; Transfection reagents; Vector; dsRNA; mCherry.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing of interests.
Figures
Similar articles
-
Inhibition of Semliki Forest virus replication with long double-stranded RNA in Aedes albopictus cells.Virus Res. 2025 Jul;357:199584. doi: 10.1016/j.virusres.2025.199584. Epub 2025 May 17. Virus Res. 2025. PMID: 40389163 Free PMC article.
-
Antiviral RNA interference responses induced by Semliki Forest virus infection of mosquito cells: characterization, origin, and frequency-dependent functions of virus-derived small interfering RNAs.J Virol. 2011 Mar;85(6):2907-17. doi: 10.1128/JVI.02052-10. Epub 2010 Dec 29. J Virol. 2011. PMID: 21191029 Free PMC article.
-
Gene silencing in tick cell lines using small interfering or long double-stranded RNA.Exp Appl Acarol. 2013 Mar;59(3):319-38. doi: 10.1007/s10493-012-9598-x. Epub 2012 Jul 7. Exp Appl Acarol. 2013. PMID: 22773071 Free PMC article.
-
Delivery of dsRNA for RNAi in insects: an overview and future directions.Insect Sci. 2013 Feb;20(1):4-14. doi: 10.1111/j.1744-7917.2012.01534.x. Epub 2012 Jul 5. Insect Sci. 2013. PMID: 23955821 Review.
-
RNA interference, arthropod-borne viruses, and mosquitoes.Virus Res. 2004 Jun 1;102(1):65-74. doi: 10.1016/j.virusres.2004.01.017. Virus Res. 2004. PMID: 15068882 Review.
References
-
- Galán-Huerta KA, Rivas-Estilla AM, Fernández-Salas I, Farfan-Ale JA, Ramos-Jiménez J. Chikungunya virus: a general overview. Medicina Universitaria. 2015;17:175–183. doi: 10.1016/j.rmu.2015.06.001. - DOI
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

