BUB1 regulates non-homologous end joining pathway to mediate radioresistance in triple-negative breast cancer
- PMID: 38863037
- PMCID: PMC11167950
- DOI: 10.1186/s13046-024-03086-9
BUB1 regulates non-homologous end joining pathway to mediate radioresistance in triple-negative breast cancer
Abstract
Background: Triple-negative breast cancer (TNBC) is a highly aggressive form of breast cancer subtype often treated with radiotherapy (RT). Due to its intrinsic heterogeneity and lack of effective targets, it is crucial to identify novel molecular targets that would increase RT efficacy. Here we demonstrate the role of BUB1 (cell cycle Ser/Thr kinase) in TNBC radioresistance and offer a novel strategy to improve TNBC treatment.
Methods: Gene expression analysis was performed to look at genes upregulated in TNBC patient samples compared to other subtypes. Cell proliferation and clonogenic survivals assays determined the IC50 of BUB1 inhibitor (BAY1816032) and radiation enhancement ratio (rER) with pharmacologic and genomic BUB1 inhibition. Mammary fat pad xenografts experiments were performed in CB17/SCID. The mechanism through which BUB1 inhibitor sensitizes TNBC cells to radiotherapy was delineated by γ-H2AX foci assays, BLRR, Immunoblotting, qPCR, CHX chase, and cell fractionation assays.
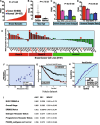
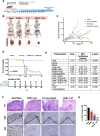
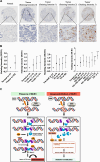
Results: BUB1 is overexpressed in BC and its expression is considerably elevated in TNBC with poor survival outcomes. Pharmacological or genomic ablation of BUB1 sensitized multiple TNBC cell lines to cell killing by radiation, although breast epithelial cells showed no radiosensitization with BUB1 inhibition. Kinase function of BUB1 is mainly accountable for this radiosensitization phenotype. BUB1 ablation also led to radiosensitization in TNBC tumor xenografts with significantly increased tumor growth delay and overall survival. Mechanistically, BUB1 ablation inhibited the repair of radiation-induced DNA double strand breaks (DSBs). BUB1 ablation stabilized phospho-DNAPKcs (S2056) following RT such that half-lives could not be estimated. In contrast, RT alone caused BUB1 stabilization, but pre-treatment with BUB1 inhibitor prevented stabilization (t1/2, ~8 h). Nuclear and chromatin-enriched fractionations illustrated an increase in recruitment of phospho- and total-DNAPK, and KAP1 to chromatin indicating that BUB1 is indispensable in the activation and recruitment of non-homologous end joining (NHEJ) proteins to DSBs. Additionally, BUB1 staining of TNBC tissue microarrays demonstrated significant correlation of BUB1 protein expression with tumor grade.
Conclusions: BUB1 ablation sensitizes TNBC cell lines and xenografts to RT and BUB1 mediated radiosensitization may occur through NHEJ. Together, these results highlight BUB1 as a novel molecular target for radiosensitization in women with TNBC.
Keywords: BUB1; DNA damage response; DNAPK; NHEJ; Radiation sensitization; TNBC.
© 2024. The Author(s).
Conflict of interest statement
SS, ST, WMC, OH, SLB, MDG, AJD, SN: No competing interests, FS: Varian Medical Systems Inc - Honorarium and travel reimbursement for lectures and talks, Varian Noona – Member of Medical Advisory Board - Honorarium (no direct conflict), BM: Research support from Varian, ViewRay, and Philips (no direct conflict), CS: Exact Sciences (paid consultant - no direct conflict), EW: Genentech research support for clinical trials.
Figures





Update of
-
BUB1 regulates non-homologous end joining pathway to mediate radioresistance in triple-negative breast cancer.bioRxiv [Preprint]. 2024 May 10:2024.05.07.592812. doi: 10.1101/2024.05.07.592812. bioRxiv. 2024. Update in: J Exp Clin Cancer Res. 2024 Jun 11;43(1):163. doi: 10.1186/s13046-024-03086-9. PMID: 38766122 Free PMC article. Updated. Preprint.
Similar articles
-
BUB1 Inhibition Sensitizes TNBC Cell Lines to Chemotherapy and Radiotherapy.Biomolecules. 2024 May 25;14(6):625. doi: 10.3390/biom14060625. Biomolecules. 2024. PMID: 38927028 Free PMC article.
-
BUB1 regulates non-homologous end joining pathway to mediate radioresistance in triple-negative breast cancer.bioRxiv [Preprint]. 2024 May 10:2024.05.07.592812. doi: 10.1101/2024.05.07.592812. bioRxiv. 2024. Update in: J Exp Clin Cancer Res. 2024 Jun 11;43(1):163. doi: 10.1186/s13046-024-03086-9. PMID: 38766122 Free PMC article. Updated. Preprint.
-
Seviteronel, a Novel CYP17 Lyase Inhibitor and Androgen Receptor Antagonist, Radiosensitizes AR-Positive Triple Negative Breast Cancer Cells.Front Endocrinol (Lausanne). 2020 Feb 11;11:35. doi: 10.3389/fendo.2020.00035. eCollection 2020. Front Endocrinol (Lausanne). 2020. PMID: 32117061 Free PMC article.
-
Enhancing radiotherapy techniques for Triple-Negative breast cancer treatment.Cancer Treat Rev. 2025 May;136:102939. doi: 10.1016/j.ctrv.2025.102939. Epub 2025 Apr 17. Cancer Treat Rev. 2025. PMID: 40286498 Review.
-
Tumor Microenvironment Dynamics of Triple-Negative Breast Cancer Under Radiation Therapy.Int J Mol Sci. 2025 Mar 20;26(6):2795. doi: 10.3390/ijms26062795. Int J Mol Sci. 2025. PMID: 40141437 Free PMC article. Review.
Cited by
-
PIPKIγ promotes non-homologous end joining through LIG4 to enhance radiotherapy resistance in triple-negative breast cancer.Cell Death Dis. 2025 Jul 31;16(1):578. doi: 10.1038/s41419-025-07894-5. Cell Death Dis. 2025. PMID: 40744919 Free PMC article.
-
Present and Future of Immunotherapy for Triple-Negative Breast Cancer.Cancers (Basel). 2024 Sep 24;16(19):3250. doi: 10.3390/cancers16193250. Cancers (Basel). 2024. PMID: 39409871 Free PMC article. Review.
-
BUB1 Inhibition Overcomes Radio- and Chemoradiation Resistance in Lung Cancer.Cancers (Basel). 2024 Sep 27;16(19):3291. doi: 10.3390/cancers16193291. Cancers (Basel). 2024. PMID: 39409911 Free PMC article.
-
MYO1B promotes radioresistance in head and neck squamous cell carcinoma by regulating tumor stemness and DNA damage repair via the PI3K/AKT pathway.Cancer Cell Int. 2025 Jul 2;25(1):248. doi: 10.1186/s12935-025-03863-2. Cancer Cell Int. 2025. PMID: 40604770 Free PMC article.
-
BUB1 Inhibition Sensitizes TNBC Cell Lines to Chemotherapy and Radiotherapy.Biomolecules. 2024 May 25;14(6):625. doi: 10.3390/biom14060625. Biomolecules. 2024. PMID: 38927028 Free PMC article.
References
-
- Kyndi M, Sorensen FB, Knudsen H, Overgaard M, Nielsen HM, Overgaard J, et al. Estrogen receptor, progesterone receptor, HER-2, and response to postmastectomy radiotherapy in high-risk breast cancer: the Danish Breast Cancer Cooperative Group. J Clin Oncol. 2008;26(9):1419–26. doi: 10.1200/JCO.2007.14.5565. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

