Downregulation of the metalloproteinases ADAM10 or ADAM17 promotes osteoclast differentiation
- PMID: 38863060
- PMCID: PMC11167776
- DOI: 10.1186/s12964-024-01690-y
Downregulation of the metalloproteinases ADAM10 or ADAM17 promotes osteoclast differentiation
Abstract
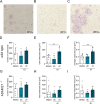
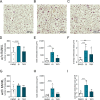
Bone resorption is driven through osteoclast differentiation by macrophage colony-stimulating factor (M-CSF) and receptor activator of nuclear factor kappa-Β ligand (RANKL). We noted that a disintegrin and metalloproteinase (ADAM) 10 and ADAM17 are downregulated at the expression level during osteoclast differentiation of the murine monocytic cell line RAW264.7 in response to RANKL. Both proteinases are well known to shed a variety of single-pass transmembrane molecules from the cell surface. We further showed that inhibitors of ADAM10 or ADAM17 promote osteoclastic differentiation and furthermore enhance the surface expression of receptors for RANKL and M-CSF on RAW264.7 cells. Using murine bone marrow-derived monocytic cells (BMDMCs), we demonstrated that a genetic deficiency of ADAM17 or its required regulator iRhom2 leads to increased osteoclast development in response to M-CSF and RANKL stimulation. Moreover, ADAM17-deficient osteoclast precursor cells express increased levels of the receptors for RANKL and M-CSF. Thus, ADAM17 negatively regulates osteoclast differentiation, most likely through shedding of these receptors. To assess the time-dependent contribution of ADAM10, we blocked this proteinase by adding a specific inhibitor on day 0 of BMDMC stimulation with M-CSF or on day 7 of subsequent stimulation with RANKL. Only ADAM10 inhibition beginning on day 7 increased the size of developing osteoclasts indicating that ADAM10 suppresses osteoclast differentiation at a later stage. Finally, we could confirm our findings in human peripheral blood mononuclear cells (PBMCs). Thus, downregulation of either ADAM10 or ADAM17 during osteoclast differentiation may represent a novel regulatory mechanism to enhance their differentiation process. Enhanced bone resorption is a critical issue in osteoporosis and is driven through osteoclast differentiation by specific osteogenic mediators. The present study demonstrated that the metalloproteinases ADAM17 and ADAM10 critically suppress osteoclast development. This was observed for a murine cell line, for isolated murine bone marrow cells and for human blood cells by either preferential inhibition of the proteinases or by gene knockout. As a possible mechanism, we studied the surface expression of critical receptors for osteogenic mediators on developing osteoclasts. Our findings revealed that the suppressive effects of ADAM17 and ADAM10 on osteoclastogenesis can be explained in part by the proteolytic cleavage of surface receptors by ADAM10 and ADAM17, which reduces the sensitivity of these cells to osteogenic mediators. We also observed that osteoclast differentiation was associated with the downregulation of ADAM10 and ADAM17, which reduced their suppressive effects. We therefore propose that this downregulation serves as a feedback loop for enhancing osteoclast development.
Keywords: Colony stimulating factor 1 receptor; Knockout mice; Metalloproteinase; Monocyte; Osteoclast; Receptor activator of nuclear factor kappa-Β ligand; Shedding.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
GPR37 is processed in the N-terminal ectodomain by ADAM10 and furin.FASEB J. 2021 Jun;35(6):e21654. doi: 10.1096/fj.202002385RR. FASEB J. 2021. PMID: 34042202 Free PMC article.
-
Regulation of podocyte surface proteins by the enzyme A Disintegrin And Metalloproteinase 10 (ADAM10).Kidney Int. 2025 Aug;108(2):214-232. doi: 10.1016/j.kint.2025.04.010. Epub 2025 May 6. Kidney Int. 2025. PMID: 40339751
-
High mobility group box 1 protein regulates osteoclastogenesis through direct actions on osteocytes and osteoclasts in vitro.J Cell Biochem. 2019 Oct;120(10):16741-16749. doi: 10.1002/jcb.28932. Epub 2019 May 20. J Cell Biochem. 2019. PMID: 31106449 Free PMC article.
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
Cited by
-
ADAM Proteases in Cancer: Biological Roles, Therapeutic Challenges, and Emerging Opportunities.Cancers (Basel). 2025 May 19;17(10):1703. doi: 10.3390/cancers17101703. Cancers (Basel). 2025. PMID: 40427200 Free PMC article. Review.
-
Identification of a key environment-responsive gene mediating environmental impact on postmenopausal osteoporosis.Front Public Health. 2025 Mar 27;13:1536851. doi: 10.3389/fpubh.2025.1536851. eCollection 2025. Front Public Health. 2025. PMID: 40213422 Free PMC article.
References
-
- Gasser JA, Kneissel M. Bone physiology and Biology. In: Smith SY, Varela A, Samadfam R, editors. Bone toxicology. Cham: Springer International Publishing; 2017. pp. 27–94.
-
- Hakozaki A, Yoda M, Tohmonda T, Furukawa M, Hikata T, Uchikawa S, Takaishi H, Matsumoto M, Chiba K, Horiuchi K, Toyama Y. Receptor activator of NF-kappaB (RANK) ligand induces ectodomain shedding of RANK in murine RAW264.7 macrophages. J Immunol. 2010;184(5):2442–8. doi: 10.4049/jimmunol.0901188. - DOI - PubMed
-
- Horiuchi K, Miyamoto T, Takaishi H, Hakozaki A, Kosaki N, Miyauchi Y, Furukawa M, Takito J, Kaneko H, Matsuzaki K, et al. Cell surface colony-stimulating factor 1 can be cleaved by TNF-alpha converting enzyme or endocytosed in a clathrin-dependent manner. J Immunol. 2007;179(10):6715–24. doi: 10.4049/jimmunol.179.10.6715. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

